Monday, February 14, 2022
Blond hair is only indirectly associated with Anatolian ancestry in Estonia...duh
In a recent paper about complex traits in Europeans, Marnetto et al. found that blond hair and blue eyes showed a relatively high association with ancient Anatolian ancestry.
This is a somewhat curious finding considering that ancient Anatolians weren't particularly blond haired or blued eyed, and that's probably an understatement.
However, the Europeans that Marnetto et al. based their analysis on were Estonians. And in Estonia ancient Anatolian ancestry peaks in the west and north, probably because this is where Estonians have the most Germanic and Finnish ancestry.
Germanic and Finnish populations are somewhat richer in ancient Anatolian ancestry than Estonians, and, unlike ancient Anatolians, they're often exceptionally blond haired and blue eyed.
So it makes sense that, in Estonia at least, ancient Anatolian ancestry is associated with blond hair and blue eyes, but only indirectly so. The more direct link is between Germanic and Finnish ancestry and blond hair and blue eyes.
I feel that Marnetto et al. should've investigated this, and they also should've made it clear that the associations they found won't necessarily be seen in other European countries.
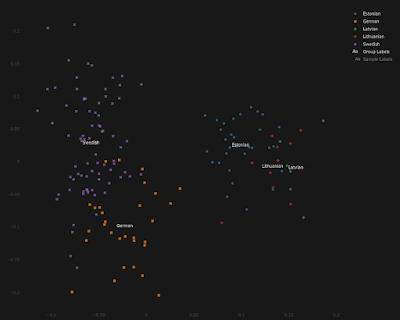
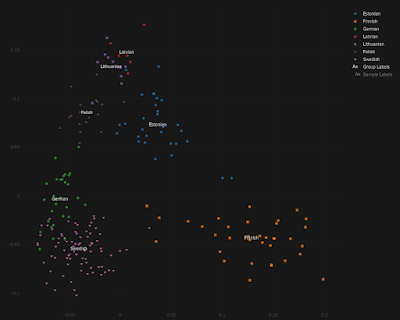
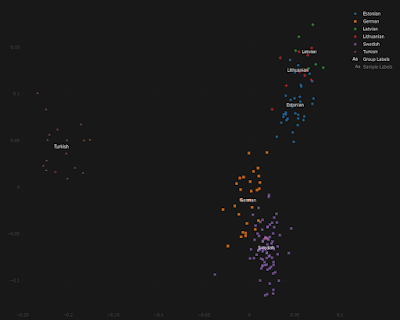
For the doubters out there, and I know there are at least a few of you, below is a series of Principal Component Analyses (PCA) showing how Estonians compare to other populations from around the Baltic Sea, as well as to present-day Turks from central Anatolia.
Note that, by and large, the same Estonians who show more affinity to the Germanic and/or Finnish individuals are also shifted slightly closer to the Turks, and this is because they harbor elevated ancient Anatolian ancestry. The relevant datasheets are available here.
Citation...
Marnetto et al., Ancestral genomic contributions to complex traits in contemporary Europeans, Current Biology (2022), https://doi.org/10.1016/j.cub.2022.01.046
See also...
Ancient ancestry and complex traits in Estonians (Marnetto et al. 2022)
Mainstream media BS: Europeans owe their height to Asian nomads
228 comments:
Read the rules before posting.
Comments by people with the nick "Unknown" are no longer allowed.
See also...
New rules for comments
Banned commentators list

I read that myself along with the mainstream media reports repeating this nonsense. Somebody on quora posted the pic and when I first saw it I thought it was a bad joke.
ReplyDeletehttps://i.dailymail.co.uk/1s/2022/02/08/16/53927901-10489451-image-a-5_1644336230066.jpg
By the way I am not really familiar with your recent work. I have used Eurogenes on GEDmatch for quite a while (it thinks I'm North German or Danish depending on the calculator). But I think all the stuff there is out of date. I see you now use G25. Is this something you developed? What is it based on exactly?
@Davidski - you wrote "Germanic and Finnish populations are somewhat richer in ancient Anatolian ancestry than Estonians, and, unlike ancient Anatolians, they're often exceptionally blond haired and blue eyed... The more direct link is between Germanic and Finnish ancestry and blond hair and blue eyes."
ReplyDeleteGiven that Estonians speak Finnic languages, would that suggest that the Finnic ancestry was there from the start? I mean, Finns and Estonians would have common ancestry, so the Anatolian part would have been in both peoples? Or that the Finns were earlier exposed to Anatolian ancestry that the Estonians were not? How did that work?
So is it that neither Finns nor Estonians tend to blonde + blue eyes and it all came from other European admixes?
thanks for looking at this
@jc1331
ReplyDeleteThe G25 is a PCA analysis. The GEDmatch calcs are Admixture tests.
https://eurogenes.blogspot.com/2019/07/getting-most-out-of-global25_12.html
@LivoniaG
ReplyDeleteMany Estonians do actually have Finnish ancestry, as opposed to just Finnic ancestry.
You can see this in IBD tests and in fine scale PCA, with many Estonians pulling heavily towards modern Finns.
Southern Finns do have more Anatolian ancestry than many Estonians, especially those that live near the Latvian border, so in my view both Germanic and southern Finnish admixture in northern Estonians is making them look a little more Anatolian-related than the national average.
Someone should do a study on this, like maybe the guys at the Estonian Biocentre.
Oh, wait...
Iranian related ancestry probably brought Indo-Iranian languages into Iran, central Asia & Northern India.
ReplyDeleteUnless someone is stupid enough to conclude that this entire belt spoke proto-Dravidian language.
Presence of Fire temples, soma technology, knowledge of horses and wagons in BMAC prior to steppe intrusion is a giveaway for this.
16 perfect lands of Ahura Mazda is where you will find Iranian rich population groups.
@AshishKaull
ReplyDeleteIranian related ancestry probably brought Indo-Iranian languages into Iran, central Asia & Northern India.
LOL
I would like to know what was the percentage of Iranian related ancestry in this region.
ReplyDeleteElamites were non-Indo-Iranian and my guess is they were Anatolian rich with minor Iranian ancestry.
IVC periphery on average was 75-80% Iranian related. If it was that high in AASI, steppe theory would have been valid but not any more.
Who were ancestors of Iran_N?
ANE + D....
Elamites must have been largely Iran N, and proto-Elamites were probably 100% Iran N.
ReplyDeleteThat's easy to work out, considering that the Iran N samples we have come from close to the proto-Elamite homeland.
Big deal of 2016: the territory of present-day Iran cannot be the Indo-European homeland
https://eurogenes.blogspot.com/2016/11/big-deal-of-2016-territory-of-present.html
3500bce & later
DeleteWestern Iran where Elamite originated has high Anatolian admixture. Eastern Iran & BMAC has smaller Anatolian admixture.
Is that so?
or Am I missing new data?
My guess is Airyanem Vaejah is where this group originated. If Indo-Iranian originated in CWC Airyanem Vaejah would have been in Europe which clearly not the case.
ReplyDeleteAiryanem Vaejah was in Turkmenistan in the Yaz culture.
ReplyDeleteIndeed, the early Iron Age Yaz II archaeological culture, located in southwestern Turan, is generally classified as an Iranian culture, and even posited to have been the Airyanem Vaejah, aka home of the Iranians, from ancient Avestan literature.
https://eurogenes.blogspot.com/2018/07/an-early-iranian-obviously.html
Eastern shores of Caspien or somewhere nearby.
ReplyDeleteOnly difference being I am linking them to an older Iranian HG related group and not to steppe related people.
Proto-Indo-Iranians came from the steppe.
ReplyDeleteAre ancestors of CHG-Iran_N-Iranian related; Dzudzuana + ANE?
DeleteIf so Proto-Indo-Iranians came from someplace between Dzudzuana Cave and Siberia.
blue eyes would be inherited from a WHG population as can be seen in the example of the girl from the island of Lolland and blonde hair was inherited in Europe from steppe people who had EHG similar input who in turn had an ANE input.
ReplyDeleteThe origin of blonde Hair is in Yana Russia KIT 18,000 ya ( ANE Population) and not ANF ! And arrive to Europe with EHGs ( ANE component )
These are the timelines some in our group are looking into:
ReplyDelete2200-2000bce BMAC (Proto-Iranian)
2200-2000bce IVC (Proto-Vedic)
2000bce +- 100 years.
Battle of 10 kings.
Migration eastwards of IVC people in places like Sanauli
1700-1500bce
Mahabharata war
400bce
Nanda dynasty
~300bce Greek-Indian interaction
BMAC can't be proto-Indo-Iranian.
ReplyDeleteIt doesn't fit the profile.
Davidski how can BMAC best model, think predominantly an Iran_CHL population and later input from Andronovo( Proto-Iranian)or rather taken over?
ReplyDeleteBMAC must be mostly derived from local Neolithic groups, like Geoksyur_N.
ReplyDeleteThere's no evidence as yet of BMAC being taken over or anything like that.
Right now, there's only really evidence of widespread mixture between post-Andronovo and post-BMAC groups.
@AshishKaull
ReplyDeleteDid the bronze age ancestors of Finnic and Saamic people also live in or around the BMAC? Because there are several words in those languages that are derived from "Arya". Lots of loanwords from Indo-Iranian and Iranic, but also coming from people who called themselves Aryan obviously.
Might wanna read the Rigveda as well because you can pick up a thing or two about subsistence economies of the Rigvedic Aryans, which is something you should have knowledge in if you want to tie in a people to a particular material culture.
As far as loans are concerned I haven't looked much into it. So it would be too early for me to make an argument related to that.
Delete2200bce Northern cities of IVC were abandoned (century long draught in Mesopotamia & Indus valley, River changing course/River drying up) and people returned to rural subsistence economy.
Vedic people are well aware of Cities/Towns which are missing from 1500bce onwards.
I have no idea about the exact nature of loans.
DeleteMy guess
Iranic loans = Scythian influence
Aryan loans could be similar to those found among Hittites and Hurrians. A small elite or trading or Priest community establishing itself in the Finnic regions.
Andronovo and BMAC agree with the formation of the predominantly Central Asian population, with different proportions according to ethnicity
ReplyDeleteFor areas where blond hair and light eyes are relatively common (such as north/western Europe) is it naive to ask whether these recessive traits arrived together at the same time, already a package in the population they came with?
ReplyDeleteSince it is perfectly feasible to have brown eyes (dominant) and fair hair — Is it worth considering when and where the recessives met up in numbers?
@AshishKaull
ReplyDeleteA small elite or trading or Priest community establishing itself in the Finnic regions.
Objectively speaking, what are the chances that this sort of bullshit is true, as opposed to Andronovo spreading Indo-Iranian languages across Asia, like its DNA?
I don't know what Andronovo spoke?
DeleteAnyway I would like to know the exact nature of Aryan loans. Which words exactly?
Are there Finnic loans into Vedic? As far as I know there are none. It appears one way borrowing.
I suppose we see one way borrowing because the Iranian dialects which interacted with Siberian languages died (except Ossetian, but they where not the northernmost).
Delete@Steppe
ReplyDeleteThanks, I was looking in the same direction. There are light skin/blue eyes/blond hair in Fennoscandia/Baltic before any EEF admixture and this can explain this variant of "blondism" and in line what Davidski explains. One curiosity I found was some central/northern Bell Beakers groups that was very "blond", was there a connection with The Nordic Bronze Age? Where else did they pick up this, not from Corded Ware or Yamnaya if I got it right?
@Wee e
One theory of origin is where Palaeolithic WHG moving north and met the EHG coming south, in central Scandinavia. WHG with blue eyes and EHG with light pigmentation.
Yes, SHG is a mixture of WHG and EHG, but the high proportion of blondes in the Bell Beakers is not known to me, rather that they are dark-haired, but BBs have their largest genetic proportion of CWC (derived culture) only more EEF and WHG, but it is best to ask Davidski . And BBs Iberia is more an exchange of ideas (cultural influence) but genetically nothing to do with the Central European BBs.
DeleteI don't know anything about exceedingly blond Bell Beakers.
ReplyDeleteSome of them may have been blond, but if so, that's not surprising because there were markers for blond hair in Corded Ware people, and Bell Beakers came from western Corded Ware.
After Corded Ware, there was some intense selection for various traits in Northern Europe, like for lactase persistence, blond hair, blue eyes, maybe height as well.
So it's not like any of the ancient populations had to be super blond to be ancestral to the blonds of Northern Europe.
@Davidski
ReplyDelete"After Corded Ware, there was some intense selection for various traits in Northern Europe, like for lactase persistence, blond hair, blue eyes, maybe height as well."
Wasn't this also for pigmentation? It seems the Yamnaya-related peoples were "swarthier" compared to modern-day northern Europeans at least.
I think the Yamnaya people had a slightly darker skin than today's Central Europeans, like today's Romanians or Tatars, or rather, a medium-light to slightly brown skin.
DeleteAre there any burnt cities in Andronovo horizon?
ReplyDeleteI find the mention of City destroying acts of Indra & Agni quite interesting.
2 known sites in your traditional Indo-Iranian speaking regions are;
2700bce
Shahr-e Sukhteh (Harappans were there)
Early Harappan phase
Kot Diji
Andronovo didn't have cities, because they were nomads, like the earliest Indo-Iranians.
ReplyDeleteBMAC had cities. See that's why BMAC can't be Indo-Iranian.
Yes. I am aware of that.
DeleteBMAC can't be proto-Indo-Iranian.
BMAC appears to be in a proto-Iranian phase or to be precise proto-Zoroastrian phase with its Fire temples and soma/Homa like features ~2000bce.
All this fits well with Iranian related ancestry as source for Indo-Iranian languages/culture not steppe.
These features post Andronovo admixture would have strengthened Andonovo = Proto-Indo Iranian.
7 samples belonging to haplogroup J & 4 belonging to R out of 18 male samples from main BMAC population.
@Davidski
ReplyDelete"The G25 is a PCA analysis. The GEDmatch calcs are Admixture tests.
https://eurogenes.blogspot.com/2019/07/getting-most-out-of-global25_12.html"
Yes I have seen this before which is why I asked about it. What I was asking for was the technical details of how they are determined. I saw a couple of companies offering these like Illustrative DNA and you after July for instance, but that gets more than awkward if you want to calculate more than a few or want something right away. Or is this propietary information? I saw one guy who was estimating these from gedmatch by using his own results there and taking the oracle results with a sheet with known g25 coefficients and estimating it that way. Not sure how accurate that would be.
I saw one guy who was estimating these from gedmatch by using his own results there and taking the oracle results with a sheet with known g25 coefficients and estimating it that way. Not sure how accurate that would be.
ReplyDeleteThat might work OK in some instances, although I doubt it would be super accurate.
I would need to run the files, but as you pointed out, not right now.
Rig Veda 7.6.5
ReplyDelete"Him who brake down the walls with deadly weapons, and gave the Mornings to anoble Husband,
Young Agni, who with conquering strength subduing the tribes of Nahus made them bring their tribute."
"There are obvious signs of extensive burns over the entire site, including both the lower habitation area and the high mound (the fortified town), which were also observed at other Early Harappan sites: Period III at Gumla, Period II at Amri, Period I at Naushero. Signs of cleavage were observed at Early Harappan phase Period I at Kalibangan. The cause of the disruptions and/or abandonment of these sites toward the end of the Early Harappan phase remains unexplained."
In Rig Veda 7.6 the composer of hymns is narrating a past event, if my understanding is correct :)
One qualifying comment about this (despite my gut feeling to agree with you) is that on the previous thread "Jorge" mentioned (though I'm not sure if he understood what they'd done) is that if I'm understanding correctly, that their genome wide association of ancient ancestry in modern Estonians had a different connection with pigment traits, compared to the main analysis, which looked specifically at whether ancestry *around* SNPs in candidate regions was enriched in particular ancient ancestry in particular ways. (They're using a technique that's like local ancestry inference).
ReplyDeleteThat may reinforce the idea that there is some complex picture impacted by selection in subpopulations and this only provides a very indirect perspective on the populations that were about 8-5 kya.
There is also the potential for sex biased admixture processes combined with sex specific trait selection to affect results (e.g. what if sex biased admixture involving EEF and CW was slightly biased to blonder females within EEF?), and have been overwritten in multiple directions. (E.g. in the Baltic we know from Saag that CWC showed sex biased EEF admixture, which since declined down to levels that are not detectable).
@All,
ReplyDeleteSince we're talking about pigmentation. This study from 2019 can help us understand the true skin color of ancient Eurasians.
Loci associated with skin pigmentation identified in African populations
https://www.science.org/doi/10.1126/science.aan8433
It identified many SNPs which differentiate skin color between Brown and Black skin.
I bet WHG had the brown skin genes. We have also known the assumption WHG, Paleolithic Eurasians had African style black skin just because they lacked the light skin SLC genes is a bad assumption.
Thanks to this study we can prove it is a bad assumption. We just have to test WHG for the SNPs they found.
The distribution of the SNPs they found is really interesting.
ReplyDeleteThe "Black" allele in all of the SNPs, is as common in Papua New Gunia as in Africa.
The "Light/brown" allele is always unanimous across Eurasia, except for in India and parts of the Middle East where some have the "Black" allele.
The "Light/Brown" allele is also always unanimous in San Africans.
This all fits perfectly with skin color we see in real life.
This suggests the evolution towards lighter skin started very early in Eurasia. The same genes were then selected again independently in Arica by the ancestors of the San.
I bet if we test these SNPs on paleolithic Europeans, we'll see they had the "Light/brown" alleles. WHG definitely did.
Keep in mind the blue eye gene also lightens skin almost as much as the classic SLC genes. The blue HERC2 gene may have been selected to lighten skin color in WHG not just eye color.
If WHG had same alleles as Ameridians in skin color SNPs, but also have blue eye gene, chances are they had lighter skin than Ameridians.
We're going to have to update reconstructions of WHG.
@Davidski
ReplyDeleteJust for the record, on the map you embedded in your post one of counties with peak Anatolian_N ancestry is Ida-Viru (Narva). That is a ~80% ethnic Slav county (mostly Russians).
I strongly disagree.
ReplyDeleteTacitus spoke in the case of Germanics of "rutilae comae" = red, golden-red, reddish yellow. You can discuss about the precise perception of Tacitus. But one thing looks clear this blond is warm blond and not ash or cool blond. Cool/ash blond can never be seen for rutilae!
Estonians have much blond hair but the percentage red haired is beneath 1%. The blonds of Estonia are overwhelming cool or ash blond.
Many of old Germanics especially around the North Sea but also in Southern Sweden carried the "rutilae comae" (MCR1) mutation see here for example:
https://docs.google.com/spreadsheets/d/15p7MiR1UIsy8ySAbTKX-ju07Zhx3cPIroEmJmedeFvw/edit#gid=0
So Germanic warm blond> Estonian ash blond, big doubts....
Then the EEF story these group is the least likely carrier of this MCR1 mutation. The other groups "Steppe" / Asian warriors ;))))) and Motala SHG did carry this mutation so there were at least more sources!
And Anatolian Farmers with MCR1 mutation had simply a problem. Ever seen a red head farmer harvesting in the burning sun (with high UV rates!)? In no time raw meat. I don't think I have to explain Davidski what the effect is of putting NW'ers in bloody hot Australia.
https://www.mirror.co.uk/news/uk-news/skin-cancer-warning-red-heads-8405760
I would have agreed on everything up until very recently: on top of presumably various Uralic-related populations (like the stock basal to Udmurts) it seems that *some* Balkano-Anatolians were very early depigmented people. So it’s more nuanced than this. Probably related to Iron Gates HGs in some manner.
ReplyDeleteI don’t get what people struggle to understand with BMAC… the Aryans established themselves as a casted chariot elite. It doesn’t mean anything with a settlement isn’t Aryan. It’s certainly not to say they’re not descended from Sintashta. But BMAC-interacting populations would naturally become much more influential than pre-Aryan populations remaining on the Steppe because of population growth and overflow (this isn’t evident with Sintashta etc, and population overflow or displacement is the single rule for motivating Steppe domination like you’d see with the Scythians. I think BMAC (or that general sphere) was the nucleus of Aryan development based on various etymologies, that Sintashta-derived populations formed the casted chariot elite in BMAC with one branch departing to other trade-rich centres earlier instead of remaining in the BMAC economic sphere (as in Indo-Aryans to India and the Mitanni ruling class to the Middle East) and later on population overflow caused the divergence of the Iranians proper.
ReplyDeleteI am arguing for arguments sake. I am probably wrong here but...
DeleteMinor Ruling elite at best can provide loans to a language. It can't displace language family.
Mitanni elites were Indo-Aryan in character but linguistically they were Hurrian speaking.
Scythians during their 400+ year rule in India used Sanskrit but it never became a common language.
Turk elite minority during their 500+ year rule used Persian as a court language. A large number of Persian loans into Aryan languages but not language displacement.
British during their 190 year rule also provided many loans to Indian languages.
Dravidian kings also patronized Sanskrit. Many sanskrit loans made its way into Dravidian languages but didn't replace those languages.
Whatever Andronovo steppe related people spoke was loaned into Indian languages. Highly unlikely it replaced language family with 20-30% steppe ancestry that too when it came in small waves instead of single major migration.
Small waves barely leave trail. A major migration will leave both genetic and archaelogical trail.
Here is another funny thing; one normal Indian conversation...
"Mere delivery kis time hogi?"
"Aaglae hafte, Sir."
(Hindi/Urdu+Persian+English)
This Gibberish of a language is attrocious. Sprachbund in effect.
There is one controversial view among certain sections of Indian scholars who claim Prakrits are older than Sanskrit.
@Matt
ReplyDeleteThat makes sense to me. Or, at least, it doesn't contradict anything.
Anatolian ancestry can still be indirectly associated with light pigmentation when it comes to global genomic ancestry.
But at local level a different type of ancient ancestry might be linked to light pigmentation, because due to selection even the people who have elevated global Anatolian ancestry might carry non-Anatolian haplotypes surrounding pigmentation genes.
@Slumbery
ReplyDeleteThe paper says this:
Conversely, as shown by median covA for the Siberian component in Figure S1I, the Siberian ancestry seems to be more abundant in northeast Estonia, consistently with Finnish ancestry shown by Pankratov and colleagues. 18
Also, as far as I know, the Estonian Biocentre calls people Estonian if they speak Estonian. So my guess is that this paper only sampled Estonian-speaking Estonians, not Russian speakers.
Moreover, the Russians in northeast Estonia are likely to be very Finnic (Karelian?) admixed anyway.
@Davidski, or the other way round (which I think was what they seem to find?), sure. The patterns are not simply interpretable with easily inferring the phenotype of the ancient population anyway.
ReplyDeleteSide note, Pagani's the lead author here and his group seems to have something of a specifically in Local Ancestry Inference around functional variants. Here's an example in Indians - https://academic.oup.com/mbe/article/36/8/1628/5364274 - "Ancestry-Specific Analyses Reveal Differential Demographic Histories and Opposite Selective Pressures in Modern South Asian Populations" - Yelmen et al 2019. In this case reconstruction of the background around the SLC45A2 variant indicates that it was probably introduced at a higher frequency by Sintashta related populations, then reduced in frequency by natural selection.
It seems to me like issues here are that admixing individuals might not be typical for their group (which might lead to an association of ancestry around the variant that was not typical for the group), and then further selection could also happen and admixture between groups under different selection. Complicated.
@Sam/Genos; without getting into whether that specific suggestion possible or not, think it might be a good guess to assume the WHG who lacked variants at SLC24A5 and SLC45A2 had a similar pigmentation to Native American groups.
For some explanation, I saw this paper late last year: https://www.biorxiv.org/content/10.1101/2021.11.24.469305v3.full.pdf
This estimates the effect size per allele of SLC24A5 and SLC45A2 at -5.8 and -2.8 "Melanin Units". Europeans today in this paper have "Melanin Index" 21 ("For comparison, the MI averaged 25 and 21 for people of East Asian and European ancestry, respectively, as measured with the same equipment in our laboratory "). Therefore we'd expect a person who was genetically European today but had two ancestral alleles of each at SLC24A5 and SLC45A2 would be predicted to be 17.2 Melanin Index darker, or MI 38.2.
This paper also has a regression line which estimates that a Kalingo (the population under study, a mixed population of primarily Native American and African ancestry) person with 100% Native American ancestry would have MI 37.5, while one with 100% African ancestry would have MI 62. That is about the range that could be expected based on reported African results (https://www.sciencedirect.com/science/article/pii/S0092867417313247 - "(I)n South Africa, the Xhosa and admixed Coloured populations, have respectively darker (M index = 67.1 ± 7.5) and similar (M index = 53.1 ± 8.5) pigmentation compared to the KhoeSan populations" and Skin color is substantially darker in equatorial Ghanaians, where M index reaches a mean of 96.04 ± 10.94.).
Assuming that WHG lacked SLC24A5 and SLC45A2 derived variants, but were otherwise had similar variations to present day Europeans (IRF derived variant and OCA2 derived "blue eyes" variant slightly higher than many Europeans today, other variants slightly lower), then would expect them to have something similar to that MI of 38.2. Closer to Native Americans (a little lower or higher) than Africans anyway.
The exact numbers could change if different equipment were used but it seems like its a reasonable bet to estimate that the WHG (Western European variant) were more similar in that trait to NA groups than African groups.
correlation and causality can be difficult to untangle, just not in this case.
ReplyDelete@matt,
ReplyDeleteThanks for the link to the study. This is a good study on skin color SNPs.
It also introduces me to Kalinago people. I didn't know a population with "no" European ancestry and mostly Amerdian ancestry lived in the Caribbean.
People were asking about skin cancer. Two major distinctions
ReplyDelete(a) non-Melanomatous skin cancer - Basal Cell & Squamous Cell. Very common, usually not too nasty. This is classic 'bad mix' of chronic sun exposure, poor ozone and 'Gaelic skin' seen in Aus & NZ
(b) Melanoma- very concerning (can metastasize, even many years after apparent cure). Thought to be due to episodes of severe burning rather than chronic dose-responce. Several different subtypes with mocelular predisposition, but no clear inheritence syndromes (apart from familial dysplastic syndrome, which accounts for small minority of incidence). Again, often its the white skin in harsh environment. But there are types with different molecular aetiology which Asians and Dark-skinned people can get (e.g back of eye, under Nails or sole of foot - i.e. nonSun-exposed regions). i.e. Bob Marley
At the very least, their results should've been corroborated with a cohort from western or southern Europe.
ReplyDeleteAnd the issue of Fennoscandian admixture as a potential confounding factor for blond hair should have been looked at as well.
Yeah, this study is good for seeing the effect of SNPs. There's a few others from Latin America like this. When you put it all together you can get an idea of the real effect of these SNPs.
ReplyDeleteHonestly I tend to think WHG had a little lighter skin than Ameridians. Due to OCA2.
This informs how I depict WHG in my youtube videos. I'm not doing black WHGs anymore.
@Matt
ReplyDeleteWhat's your opinion about the inferred association between Siberian ancestry and green eyes?
That seems like the least expected outcome to me.
A genomic snapshot of demographic and cultural dynamism in Upper Mesopotamia
ReplyDeletehttps://www.biorxiv.org/content/10.1101/2022.01.31.478487v1
Don't know if this was already mentioned in a previous thread but I thought it was interesting. Apparently, pretty much all Anatolian farmers had Neolithic Levantine ancestry. There was a discussion on Anthrogenica not too long about African ancestry showing up in Neolithic Anatolians as well (which Rob seems to be backing up, and Chad has mentioned the same), and by extension EEF, which would mean all West Eurasians are essentially part African. This paper doesn't bring up that angle, but it seems to strongly corroborate Levantine > Anatolian flow, which would by definition entail African > Anatolian > EEF flow too. Another interesting point that someone from AG mentioned was that Zlaty Kun is pretty much equally related to all Eurasians, except for those with "ANA" (the African side of Iberomaurusian) ancestry.
Anyone have any thoughts? I believe with the IBM samples, all the G25 attempts to model them as a mix of any Eurasian+African population seem to be terrible, which makes suspect that their "African" ancestry is not so straightforward, but I've seen qpAdm models that are apparently more successful.
@Cy Tolliver
ReplyDeleteWhy African, and not something Natufian-related?
@David
ReplyDeleteWell, Natufians would be part African by way of their IBM-related ancestry (their E1b1b being the tell. E-M78, the forefather of E-V13 has also been found in Neolithic Anatolia so I think the progression from North Africa > Levant > Anatolia > Europe shouldn't be too controversial). I know in the past direct D-stats failed to show a strong African signal in Natufians but that just seems like a quirk of that particular program, qpGraph, qpAdm seem to be able to pick it up, and again based on y-DNA it makes intuitive sense. I think there's even a Natufian-ish or African-ish signal in Iran_N, I can dig up the threads from AG if anyone is interested.
I know in the past you've said you're not super keen on exploring deep ancestry because you think it's too speculative, and that's a fair take, but it seems clear there's some unusual ancient structure from North Africa all the way east to at least Iran, whether that's African related, Basal Eurasian, something else, who knows, but I think it's a question worth exploring and I really, really hope we get a lot more aDNA from that whole area in the next few years.
It would be some form of North African mediated via LUP Levantines. Natufians themselves are too late and presently poor coverage
ReplyDelete@jc1331
ReplyDeleteI'll have to cut you off here, because this thread isn't about the G25.
I'll be making some announcements in regards to the G25 and other related stuff in July.
@Rob
ReplyDeleteWhat exactly is the "North African" component though? Is it something clearly related to more normal SSA type of ancestries, or is it perhaps just a less drifted form of Eurasian ancestry (a more pristine Basal Eurasian, I guess?) that looks superficially African?
@Rob
ReplyDeleteThis is something I’ve thought for a while now. “Basal Eurasian” is really just Ancestral North African/Ancient East African ancestry. I posted on both Anthrogenica and Eupedia, of there being AEA/ANA in EEF/ANF, a few years ago. Like Y-DNA K and mtDNA C in IUP descended Eurasians, this ANA/AEA ancestry explains the shared Y-DNA D, E, A, and mtDNA L subclades in both Eurasians and Africans. However, “African” is pretty vague terminology; it’s almost as descriptive as Eurasian. It seems to be geographically correct, but it’s only an ambiguous stand in for the North/East African Upper Paleolithic and Mesolithic genomes we currently lack. I’m curious to see what human phylogeny will look like in Mesolithic and Paleolithic Africa.
People thinking “BMAC wasn’t taken over, ipso facto no Aryans” goes against everything the Bronze Age proper shows us. If it weren’t for the presence of Aryan names, who would ever think the Mitanni were related? They had chariots but so did BMAC of the same period. It isn’t that BMAC = Aryan, but that Aryans would have had a presence later on. Trade was the name of the game, and BMAC would have been the first true trading sphere the Sintashta-derived populations encountered.
ReplyDeleteCould anybody tell me the flaw here:
pre-Aryan folk move into BMAC territory as a chariot elite seeking riches. Likewise groups splintered off from here in the same quest: one to South Asia and one to West Asia = Indo-Aryans and Mitanni Aryans. Iranian-Aryans (or Aryans proper if you like) then expand from the BMAC culture as a result of population overflow. One group goes into Iran to form the West Iranian branch and the other spills onto the Steppe replacing other similar cultures brought to extinction (call them paleo-Aryans maybe) as the East Iranian branch. The remaining branch would be the Avestan Iranian branch: the one which held control of trade enough not to need to expand with population overflow. Why does nobody consider the idea that Aryans initially were the dominant Steppe culture involved in go-between trade with the metal-producing Urals (paleo-Aryans) and BMAC proper (call them Oxans maybe)?
Basal Eurasian is pretty obviously the Back to Africa branch of mtDNA L3 and related populations.
ReplyDeleteWhat is the general consensus among the scholars about the identity of Ahura Mazda?
ReplyDeleteAssurbanipals clay tablets attest "Assara Mazaš" ~650bce about a century and a half prior to Darius old Persian attestation.
Location of Behistun Inscription appears to be near the Assyrian territory.
@Rob 'People were asking about skin cancer.'
ReplyDeleteIt's not only skin cancer, people with a ruddy kind of complex (= MCR1) are in the full sun (with high UV rate) in no time red meat (and the tan ability is lower to very low). So for Anatolian Farmers not a favorable feature.
The core is: "blond is not blond". Warm/ ruddy blond is not cool/ash blond. The "rutilae comae" = red, golden-red, reddish yellow is according to already Tacitius significant within the Germanics.
Does this make sense? Cremation was the norm in LBA Urnfield and Jastorf nevertheless we got some samples from the Lichtenstein Cave in lower Saxony Germany (LBA) and approximately 17% of them had probably red hair. Taken in account that some (like me) have the indices for red hair but still have (warm) blond hair (even in 50% of R160W cases) sometimes with light touch red (= rutilae comae).
In Estonia the amount of red hair is beneath 1%, ash blond is prevailing, so I don't see a 1:1 connection between Germanic influence (warm blond) on the Estonians (ash blond). Apples and oranges.
@ Simon
ReplyDeleteI don’t think there is a singular population of “basal eurasians”
It probably represents ancestry streams shared by certain Near Eastern populations during the late paleoluthic, with sometimes deeply divergent origins
@Davidski, not sure, I don't really understand anything at all the independent genetic basis of green eyes. Would say that both eyecolour dark and eyecolour green are associated with Siberian ancestry in the CR (candidate region) covA, and also in teh Genome-Wide association. The eyecolour light is most strongly negatively associated with Siberian in the CR (candidate region) covA (which is at least not too surprising?).
ReplyDeleteThere was some research that indicated that East Asian groups do have some variants that lighten and darken eyes (despite the lack of the major effect European variants that lead to blue eye colour) - https://www.science.org/doi/10.1126/sciadv.abd1239 - "We find evidence for genes involved in melanin pigmentation, but we also find associations with genes involved in iris morphology and structure. Further analyses in 1636 Asian participants from two populations suggest that iris pigmentation variation in Asians is genetically similar to Europeans, albeit with smaller effect sizes."
So it's possible that Siberian groups really did contribute some variant that when driven to higher frequency and/or on a European genetic background, combined with whatever selection may have happened after admixture, lead to green eyes (rather than blue eyes). (Along with disproportionately contributing variants that lead to darker eyes?). Some interaction with the other >95% of Estonian genetic background. I don't think it's saying necessarily that overall the Siberian ancestors are predicted to have green eyes anyway, or at least we can't interpret it that way.
At least in that case they can actually confirm and compare directly with what variants are present in groups with Siberian ancestry in the present day without limitation of adna. (Though these groups often have small population sizes and strong drift, at least they are living communities that can choose to share their dna with us).
@james roper
ReplyDeleteI agree with you that BMAC region was already indo iranian speaking much before the bronze age (if that is what you meant).
However, the BMAC ancestry itself has limited impact in modern Indians or even Roopkund 850CE samples. Where BMAC does have a significant impact is the swat valley iron age, but there is no evidence yet of the ancestry ingressing deeper into Indian subcontinent.
@cy_tolliver
ReplyDelete" I think there's even a Natufian-ish or African-ish signal in Iran_N"
Yes there 'basal eurasian' in CHG and Iran_N. in my modeling this is different from Iberomaurisian or Natufian or whatever else.
Its basal because its ancestral to the split between East and west eurasians. It gives about 5-15% ancestry to CHG and 20-30% ancestry to IranN.
From the paper "The genetic prehistory of the Baltic Sea region", ;
ReplyDelete"Similar to other European Mesolithic hunter-gatherers, our Baltic foragers carry a high frequency of the derived HERC2 allele which codes for light iris colour, and like SHG and EHG they already possess an increased frequency of the derived alleles for SLC45A2 and SLC24A5, coding for lighter skin colour"
With a deep analysis, is it possible to find out if it is the same variants in the Baltic region, Scandinavia and present in modern populations?
The initial EHG route to Scandinavia is very north at latitudes above 66˚ and very "artic". Later CW and Yamnaya do not carry the "blond package" at the same levels, if I got it right. And that got me to think in terms of natural selection and the routes of EHG to the region.
I do not get the explanation of sexual selection, just to say that "Blonds have more fun" doesn't really explain anything.
Grate if someone in genetics could comment on this.
@Cy Tolliver
ReplyDelete"There was a discussion on Anthrogenica not too long about African ancestry showing up in Neolithic Anatolians as well (which Rob seems to be backing up, and Chad has mentioned the same), and by extension EEF, which would mean all West Eurasians are essentially part African. "
All West Eurasians are part African and all Africans are part Eurasian and so on, this is probably the case and imo doesn't mean much. Chad here said that Anatolia_HG are part African and than he sees an Onge wave in Boncuklu, imo that's not very realistic model but then again I have no experience with qpGraph.
Lazaridis modelled Natufians as part Taforalt and by extension Anatolia_N as part Natufian, but in both cases the admixture is low, that is about 15% of the ANA component in Natufians (Assuming their qpGraph and qpAdm models are accurate) and 15% of that in Anatolia_N, that should make about 2% of ANA in Anatolia_N, hardly surprising once you consider some EEF carry E-M78.
From what I know most formal stats find Eurasians similarly related to Africans so if there was substantial African-related admixture in Eurasia how come nobody picked up in the high coverage samples?
@Rob
ReplyDelete"It probably represents ancestry streams shared by certain Near Eastern populations during the late paleoluthic, with sometimes deeply divergent origins"
What are your graphs showing about that? Would be interested. But I don't think they are that divergent, I mean look at their uniparentals, there are usually pre-IJK branches but nothing too wild, except in Taforalt most seem to post-date the "OOA bottleneck".
Subclades of E have hardly been found in any Iranian-related sample yet it was ubiquitous in Taforalt. Same for mtDNA U6 and M1.
@Simon Stevin
"Like Y-DNA K and mtDNA C in IUP descended Eurasians, this ANA/AEA ancestry explains the shared Y-DNA D, E, A, and mtDNA L subclades in both Eurasians and Africans"
So you mean to say Tibetans and Onge have Ancient East African ancestry? And which ancient Eurasian has mtDNA L subclades that don't belong to L3 of which all Eurasian mtDNA haplogroups are descended from? Ydna A hasn't been found in any ancient remains yet either (except maybe in BA Spain can't recall rn).
If there really is this East African admixture across Eurasian then fine, but so far it seems either extremely ancient (before 45kya, in this case we might just as well talk about repeated waves from Africa during the OOA event) or not the case.
Again, ANA in EEF I can see, but "Ancient East African" gene flow as far as Japan is just not parsimonious imo.
This comment has been removed by the author.
ReplyDelete@Cy Tolliver
ReplyDeleteHave to post one (tagged the wrong person) more time because this is actually intriguing "Another interesting point that someone from AG mentioned was that Zlaty Kun is pretty much equally related to all Eurasians, except for those with "ANA" (the African side of Iberomaurusian) ancestry."
Which populations did that person say have ANA/are less related to Zlaty Kun?
@weure, still an interesting discussion; I think the thing I would say is to remember is that people with one copy of the redhaired variants of MC1R do not necessarily have pale to ruddy skin.
ReplyDeleteAs you will know, you generally need to be a homozygote for the derived variant to express that phenotype, which would not be the case if it was in some population at a low frequency.
https://www.nature.com/articles/s41467-018-07691-z - (W)e find (in a large UK Biobank study) that the proportion of red-haired individuals with two MC1R alleles is 92%, whilst only 6.3% carry a single allele (despite the fact that heterozygotes are themselves *far* more common in the general population!).
Secondly other variants like the modern day European variant of SLC45A2 and IRF4 also contribute to the expression of that phenotype being the typical Northern European redhead with intensely red hair *and* very fair skin.
That all being the case, the selective pressures that were present in Anatolia could have been sufficient to keep a variant about at low frequency (if it were preferred for some reason), to then be exposed to selection in Europe.
Consider that the MC1R risks variants V60L, R151C, R160W (which you mention) that are associated with melanoma / red hair are still about in Near Eastern groups today at frequencies that are often about 50% or more of the frequency in Europeans - https://www.biorxiv.org/content/10.1101/255117v1.full (See Fig 4). We don't have to speculate at whether these variants could survive in groups in the Near East - we know that they do (in proportions that seem in excess of what could be predicted by recent shared ancestry with Europeans via Yamnaya/Steppe_EMBA/Steppe_MLBA).
If these variants are just circulating at a low kind of frequency and generally not found together as homozygotes, on top of a lower level of some other derived pigmentation traits, they're not necessarily going to be removed by selection, especially if they are slightly favoured due to some local preference for unusual colour variation or something like this.
@weure, even in Southern Spain, where the sun is quite intense, there is some evidence for positive selection on the V60L variant - https://academic.oup.com/mbe/article/30/12/2654/1014645 - "(O)ur results indicate that two selective forces shape human MC1R diversity in Sothern Europe. Similarly to Africa, purifying selection is strong maintaining the ancestral allele in Sothern Europe. But simultaneously, positive selection is responsible for an increased frequency of the V60L allele, a low-penetrance “red hair and fair skin” allele (also associated to blonde hair; Box et al. 1997). The fact that V60L is under positive selection constitutes evidence that MC1R is also playing an active adaptive role in human skin depigmentation, at least for some areas of south Europe. . Despite V60L raising melanoma risk. (Much more interesting discussion at the link!)
ReplyDeleteThere is also some complexity *within* different variants at MC1R with the degree of "penetrance" (i.e. how likely a person with them is to have red hair). The V60L allele is particularly dependent on being present in combination with other MC1R variants (and possibly othe genetic variation). See the Figure 2 - https://www.nature.com/articles/s41467-018-07691-z. This may explain why V60L is being positively selected in Spain while many other red hair related MC1R variants have been reduced in frequency by selection. (On that note, even though one gene explains red-hair so we can probably more easily work it out for ancient people, we may still need multiple sites at that gene or a means of imputing them to predict ancient people properly...)
(Derived European variant of SLC45A2 is also associated with melanoma, but still got selected for in Spain - https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0104367 - "Selection tests suggest that allele 374F is being positively selected in South Europeans, thus indicating that depigmentation is an adaptive process. Interestingly, by genotyping 119 melanoma samples, we show that this variant is also associated with an increased susceptibility to melanoma in our populations.").
Variants are actually quite likely to arise at the MC1R gene for specific reasons (https://www.thelancet.com/journals/lanchi/article/PIIS2352-4642(19)30026-4/fulltext - "The gene for the melanocortin 1 receptor (MC1R) is unusual. It is a single exon, and many mutations lead to a reliable change in whole organism pigmentation, affecting appearance and response to ultraviolet (UV) radiation. As a result, MC1R variation commonly arises as animals move into different environments and is often central to speciation."). So it's quite possible that these variants arose quite deeply in the Out-of-Africa history and were present at some low level in many West Eurasian populations by 10,000 years ago. The variants that were subject to selection in Europe could have come from anywhere.
@ Vasistha
ReplyDelete''Yes there 'basal eurasian' in CHG and Iran_N. in my modelling''
your models are full of made-up Ghost populations because they don;t adequately contain ancient references
Blogger Davidski said...
ReplyDeleteI don't know anything about exceedingly blond Bell Beakers.
I bet he has this in mind:
https://eurogenes.blogspot.com/2018/06/guest-post-we-owe-many-of-our-genetic.html
This comment has been removed by the author.
ReplyDeleteWell it does prove that more steppe ancestry doesn't necessarily mean lighter features, doesn't it?
ReplyDeleteBoncuklu_N is Pinarbasi + Iran
ReplyDeleteBarcinN is Pinarbasi + Iran + Levant_PPN
Using qpAdm, I see no need for Natufian or Taforalt or anything else here.
@Matt
ReplyDelete"I don't really understand anything at all the independent genetic basis of green eyes. Would say that both eyecolour dark and eyecolour green are associated with Siberian ancestry in the CR (candidate region) covA, and also in teh Genome-Wide association. The eyecolour light is most strongly negatively associated with Siberian in the CR (candidate region) covA (which is at least not too surprising?)."
Green eyes means very high pheomelanin! Pheomelanin is responsible for the ruddy complex from warm blond to red hair, and from chestnut/amber (bordering brown) eyes to green eyes (bordering blue)..
It's complex. But regarding the three founding populations EEF may have transmitted but I guess very marginal, because the pheomalin amount has consequences in terms of sun burns, bad tanning, skin cancer etc. ANE and EHG/SHG could be better candidates for the pheomelanin input. Because the conditions in their heartland was not signed by high UV etc.
In Lower Saxony Germany LBA (1000 BC) the situation was very intermediate with regard to pigmentation, with for certain an amount of the ruddy (pheomelanin) complex was there for sure.
See this scheme from a dissertation about the Lichtenstein Cave LBA Lower Saxony:
https://postimg.cc/hJSQyRKJ
There is nothing surprising in those Çayonu samples from South Turkey. They are what they should be for a population who lived south of Taurus mountain range and was not affected by those WHG-ish influences that seems to be present all over plain Anatolia _north_ of Taurus.
ReplyDeleteThis type of ancestry was present in Near East since very old times. It probably directly stems from the first Out of Africa pop.
So I don't see any reason why it should have an extra recent African ancestry especially given their uniparental markers. Y dna J2 and mtdna K.
I don't know was this paper discussed here. An upper Paleolithic sample from West Georgia labeled SAT29 very similar to Dzudzuana.
ReplyDeleteClosest pops in f3 stat are in Europe not in Near East just because it shares alleles with WHG and Anatolia N.
I guess this is the main reason why CHG do not work well for Steppe Piedmont.
That's because the true proto-CHG was from modern Azerbaijan and it directly moved to North while another branch moved to West Georgia mixing with those SAT29 pops giving the actual CHG we have, which is not the exact source of CHG in Steppe Piedmont.
It's possible that WHG-ish affiliation both in West Georgia and Anatolia HG have the same source.
But I am not sure of that
https://www.cell.com/current-biology/fulltext/S0960-9822(21)00818-6
We can also use the example of Georgians. Anatolian admixture has the highest in Western Georgians than Eastern Georgians. Western Georgians also do not have WHG, but light-colored hair and eyes are more common in Western Georgians than in Eastern Georgians.
ReplyDeleteReturning to the main subject.
ReplyDeleteThat picture circulates all over Facebook and it's confuse many people.
People really start to believe that heart rate and BMI is determined by genes. Dozens of non-genetic factors influences those things.
Even if there is some genetic predisposition then once more dozens genes are involved.
Just quick search gives this paper. Keep in mind that they discuss only few aspects of heart rate.
------
In the current review, we found a total of 10 genes associated with the acute heart rate response to exercise in candidate gene studies. Only one gene (CHRM2), related to heart rate recovery, was replicated in recent GWASs. Additional 17 candidate causal genes were identified for heart rate increase and 26 for heart rate recovery in these GWASs.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6529381/
@rob
ReplyDeleteOnus is on you to prove that some taforalt or natufian or whatever population contributed to CHG or IranN. So far I have not seen anything from you, apart from proclamations.
As far as I am concerned, the input to CHG and IranN is basal to the split between east and west Eurasians defined by Kostenki and Tianyuan/Onge respectively. I have made qpGraph models before with Natufian and Taforalt in them, saw no indication of what you're claiming.
@davidski
ReplyDelete"Elamites must have been largely Iran N, and proto-Elamites were probably 100% Iran N."
Lmao, I bet 1000$ that this wont be the case lol (100% IranN). Shows how poorly you know the region.
@All “ Lazaridis modelled Natufians as part Taforalt and by extension Anatolia_N as part Natufian, but in both cases the admixture is low, that is about 15% of the ANA component in Natufians (Assuming their qpGraph and qpAdm models are accurate) and 15% of that in Anatolia_N, that should make about 2% of ANA in Anatolia_N, hardly surprising once you consider some EEF carry E-M78.
ReplyDeleteFrom what I know most formal stats find Eurasians similarly related to Africans so if there was substantial African-related admixture in Eurasia how come nobody picked up in the high coverage samples?”
What’s the exact relationship between Natufians and Anatolian farmers?
On one hand, Anatolia Neolithic as in Barcin samples have Natufian admixture introgression in them. But also, I read a study that Pinatbasi was basically (Anatolian HG) a Natufian pop from UP which was later invaded by a WHG source like Iron Gates, and there’s an equidistance between WHG and Natufians.
But additionally, Levant_N was created from an admixture of almost 1:1 Natufians and Anatolian_N.
Therefore, pre-Islamic Near East, Armenians and pre-Bronze Age Europeans were pretty much not that divergent. Am I correct?
Perhaps Genos and Matt can help me here :)
As far as the Estonian paper goes, I do not know why people are so butthurt about the paper reporting a correlation (and never claiming causation) between anatolian and hair/eyes whatever.
ReplyDeleteMuch worse papers have been written. At least what is written in this one seems to be fact rather than narrative as is done in the case of south Asia. In that region, correlation always implies causation.
Speaking of West Africans - I have read that the ancestors of African-Americans were created from 2 populations: Proto-Niger-Congo originated in the so-called “Green Sahara”, and migrated south during deccication; they weren’t “black” per se as we know it but were actually more like Taforalt Ancient North Africans, or “Caspians”, which may explain their Eurasian affiliation due to some back migration to Africa 45k ybp.
ReplyDeleteThat pop who was using iron tools was mixing progressively with native West African Hunter Gatherers, who were divergent from the Baka/Mbutu/Pugmies of Central Africa. The outcome was that modern West Africans speak languages and have height of these partially-Eurasian Green Saharan but the facial features and phenotype of the original Aterian HG forager dwellers of the Niger River.
Does it make any sense? Can anyone correct me or elaborate on that?
Blogger DragonHermit said...
ReplyDeleteWell it does prove that more steppe ancestry doesn't necessarily mean lighter features, doesn't it?
If you mean that 2018 blog post, yes it does not.
It shows the thing that David mentioned up here. That all these features (blond hair, blue eyes and lactose tolerance) exploded into space after the mix between steppe and farmer (possibly because of some kind of selection). The steppe/farmer mixes are lighter than pure steppe or pure farmer.
But it also shows both had these features. The farmers more than Steppe. But both do not come close to how much the mix between Steppe and Farmer had.
@CeRcVa
ReplyDeleteThanks for the input, can you please tell us what admixture caused you to end up without half a brain and with a heavy depigmentation fetish that your underage brain loves to cling onto so much. Thanks in advance for the answer and never ever leave your plebbit containment area again, you national embarrassment.
Lol. Serously, What?
DeleteBlonde hair, red hair, light skin arose in EHG rich populations. While highly detailed data is lacking, there is some to suggest it, as well as modern populations who originated from EHG rich populations happen to carry these traits.
ReplyDelete@Matt I think the thing I would say is to remember is that people with one copy of the redhaired variants of MC1R do not necessarily have pale to ruddy skin.
ReplyDeleteI know myself as example, FTDNA:
RS1805008 CT= TC = 1 risk
RS 885479 AG= GA= 1 risk
RS 2228479 GG=AA= 2 risk
RS 1805006 CC= AA= 2 risk
RS 1110400 TT= CC= 2 risk
RS 1805005 GG= TT= 2 risk
I have warm blond hair, chestnut eyes and the skin is (just a guess) even a skin type III/ bit leaning to II Nevertheless the skin still looks ruddy no doubt.
So yes indeed, and I don't exclude southern europe but I guess this are not the most favorable circumstances for having much pheomelanine. Bigger chance of breeding out, imo. In the foggy and froggy area around the North Sea with much much lower UV rates this is much more favorable.
So yes can play soccer with 40 C degrees and this will be done, but with 21 C degrees it's more suitable.
I'm still not done with the warm (high pheomelanine) vs cool features... I guess it's still underestimated in the for example the divers kind of blonde!
@ gamerz
ReplyDelete''What are your graphs showing about that? Would be interested. But I don't think they are that divergent''
definitely some divergent ancestries make up what might be considered 'basal West Asian populations' of the Epipaleolithic, ranging from European to some form of ancestral north African
Quite simply, there is no such population as 'basal Eurasian'. It is made-up
''Subclades of E have hardly been found in any Iranian-related sample yet it was ubiquitous in Taforalt. Same for mtDNA U6 and M1.''
What's the point ? Hg E =/= basal
This comment has been removed by the author.
ReplyDelete@weure, I thought your comment about phaeomelanin being linked to green eyes was interesting.
ReplyDeleteFrom a limited Google I couldn't find anything that directly supported it (you may know of a good reference!), *but* I did see an article that might confirm your explanation or intuition - https://www.nature.com/articles/s41467-018-08147-0 - in that it shows that green eyes are a combination of a shift away from black iris colour towards light brown that is basically a > red shift and a < blue shift, and a shift towards overall lightness and less pigment. So that seems plausibly linked to a shift towards > phaeomelanin expression in the eyes as well as overall < melanin production/retention?
On the other hand, it seems to be genetically separate from the shifts caused by MC1R as the same paper finds no influence of MC1R variants on eye colour at all.
Some people in East Asia have light brown eyes or even hazel green eyes (E.g. this actess https://jpop.fandom.com/wiki/Hashimoto_Kanna?file=Kanna_hashimoto_profile.jpg), so it might be plausible that some North Asian group with many of these variants contributed an excess of them to Estonian populations in a way linked to green eye colour... as well as contributing variants linked to dark eye colour.
They state when it comes to eyes that:" For example, GWAS carried out in Europeans have mostly focused on variation in the brown to blue color spectrum. By contrast, the C (saturation) color component examined here, with which two new loci have been associated, captures variation within brown eyes (Fig. 2b) and the index SNPs at these loci have highest derived allele frequencies in East Asians (Fig. 4).". (Not that even if it's plausible that Estonians have an excess of Siberian ancestry around these variants this would mean that all Europeans with lighter brown or green eye colour would necessarily get the variants that cause this from that ancestry as related variants would be present and dispersed from a deep period of time).
I think I got a definitive clue about that paper. High pheomelanin, the yellows-red pigment, is caused by MC1R.
ReplyDeleteMelanocyte cells contain a protein called the melanocortin 1 receptor. This protein sits on the surface of melanocyte cells. The code for this protein is on the MC1R gene. When the protein is activated, it specifically produces pheomelanin. Redheads have a genetic variant of the MC1R gene that causes their melanocytes to primarily produce pheomelanin.
Indeed and this is not only the case for red hair, it is also at stake in the case of warm blond hair, green (= very big amount pheomelanine!), amber and chestnut eyes.
In the recent paper about complex traits in Europeans green eyes are negative correlated with EEF and positive with Siberian (ANE) founding population.
May be some depigmentation is correlated with EEF (Herc 2 etc). Nevertheless the negative correlation between green eyes and EEF and positive sign for ANE is imo significant.
Does it hint that the "pheomelanin pigmentation complex" is correlated with ANE? Is it a coincidence that Udmurt, the Russian people which seems to have with 25-28% the highest ANE amount (in good old ANE K8), and in an old blog of Davdski he states this people had the "highest Yamna amount" (2015) also got the worlds highest amount of red hair? No coincidence to me....
@AWood “ Blonde hair, red hair, light skin arose in EHG rich populations. While highly detailed data is lacking, there is some to suggest it, as well as modern populations who originated from EHG rich populations happen to carry these traits.”
ReplyDeleteBut what about *CHG* in Steppe?
The CHG are ANE rich (1/3 ANE + 2/3 Dzudzuana like), but no one mentions them here in that regard?
This comment has been removed by the author.
ReplyDelete@Vasistha
ReplyDeleteYour models about Boncuklu and Barcin imo agree with the affinities they have to Europe and the Levant as well as the Near East, Boncuklu for example is clearly both closer to WHG and to CHG.
@Rob
ReplyDelete"definitely some divergent ancestries make up what might be considered 'basal West Asian populations' of the Epipaleolithic, ranging from European to some form of ancestral north African
Quite simply, there is no such population as 'basal Eurasian'. It is made-up"
Basal Eurasian being made up is quite plausible, but I only wish you elaborated a bit more on what you seeing. For example, how far north does this ancestral North African influence extent to, and how recent do you think it is in the region? Is the rest "West Eurasian" or are there other influences beyond the ANE/ENA admixture in the Mesolithic?
The reason I brought up E, is because all populations inferred so far to have ANA/Taforalt-like admixture do show subclades of said haplogroups along with distinctive mtDNA lineages. That isn't really seen in samples to the east of Iran for example, ancient or modern. Never argued E=Basal, I am arguing E=ANA/Taforalt-related signal.
From the (very) little I've read of archaeology the only real movement from Africa (North Africa) to the Near East seems to be the Mushabian culture which interacted with the Kebaran to form Natufians, or so it assumed. There are prior movements from Europe to the Levant before that as well.
I am only skeptical of (non-trace amounts of) "ANA" in CHG and Iran_N that's all, it's most likely there in Natufians and some trickled down to EEF eventually.
@Andrzejewski
ReplyDeleteI would say that the Iran_Neo founding population was certainly black or brown haired with predominantly brown eyes, not sure what the implications are for the ancestral CHG population are, but I would expect much of the same. All of these ancient civilizations were tawny skinned with black hair, as per their depictions, it's just kind of funny that we need to pretend things like blonde hair and even light skin actually come from them.
From the data, it looks like somewhere on the WHG-EHG cline did the light skin and (European) light hair mutations pop up. Presumably other independent mutations for light hair have popped up in other populations, but are considerably rarer. Do we have any specimens with skin lightening earlier than the CHG one? I gather it's only a matter of time that some EHG-rich or proto-EHG rich eastern Euro pops up with the mutation.
@Andrejewski
ReplyDeleteI've already typed two messages and don't mean to spam the thread but since you "quoted" me, I'd say that the recent DATES paper kinda argued against Natufian admixture in Iron_Gates_HG. It seems Natufians just share a lot of common ancestry with Anatolia_HG (but the latter apparently lacking the ANA signal found in Natufians) so hence why the models are working I am assuming. I'd bet that Dudzuana and Iron_Gates_HG share a lot of ancestry too, perhaps the WHG -like component in Dudzuana (and as a result in Natufians etc) comes from Europe, but it could also be the reverse.
"but the facial features and phenotype of the original Aterian HG forager dwellers of the Niger River."
I don't think Aterians contributed to anybody they are too ancient to be relevant imo. If they did it's in really low amounts.
"Therefore, pre-Islamic Near East, Armenians and pre-Bronze Age Europeans were pretty much not that divergent. Am I correct?"
It's still the case, Europeans and Near Easterners are quite close today too. But yeah it seems Anatolia_N and Levant_N were indeed a lot more related to each other (phylogenetically speaking) than modern-day Europeans and modern-day Levantines. Could be wrong ofc.
This comment has been removed by the author.
ReplyDelete@ gamerz_J
ReplyDelete''The reason I brought up E, is because all populations inferred so far to have ANA/Taforalt-like admixture do show subclades of said haplogroups along with distinctive mtDNA lineages. That isn't really seen in samples to the east of Iran for example, ancient or modern. Never argued E=Basal, I am arguing E=ANA/Taforalt-related signal.''
The correlation between Y-DNA and G-w Signals, or even with linguistics for that matter, is during the 'range expansion' phase. i.e. when populations are expanding/ migrating relatively rapidly. However, this correlation can be lost when there is a secondary expansion and when there is a steady-state cross-cultural interaction zone. For ex; outside EE, the highest levels of EHG are in SHGs which wholly lack paleo-Siberian male lineages (they are all Hg I2 + a single pre-I1) whilst the R1b-rich Iron Gates or Latvian HGs have much lower levels of EHG admixture.
Thus lack of hg E in current Iran N samples is not an issue because (i) they might have acquired north African admixture from Natufians rather than directly from north Africa, and (ii) it is only a level of 20% ('only', but still rather significant). Also, this is marginally higher than in CHG, and much higher than AHG (which are ~ 0%)
@Gamerz_J
ReplyDelete"Which populations did that person say have ANA/are less related to Zlaty Kun?"
I don't remember if they specified, but at a minimum I assume IBM and Natufian/Levant_N, and I think also Iran_N and Anatalian_N too.
Some other links to ENA, and the hotspots of "pheomelanin pigmentation complex" :
ReplyDeleteKyrgyz people:
Rachel Lung, Rachel (2011). Interpreters in Early Imperial China. During the reign period of Kaiyuan of [emperor] Xuanzong, Ge Jiayun, composed A Record of the Western Regions, in which he said "the people of the Jiankun state all have red hair and green eyes. The ones with dark eyes were descendants of [the Chinese general] Li Ling [who was captured by the Xiongnu]...of Tiele tribe and called themselves Hegu."
And the Tarin Mummies seem to have had tremendous high loads of the pheomelanin complex:
http://redhairmyths.blogspot.com/202...m-mummies.html
Davidski 2015:
ReplyDelete"Now, qpAdm is easy to run but very difficult to use correctly. However, even when fumbling around like a drunk with this software, it's easy to pick up some useful hints. Clearly, even if the ancestry proportions are way off, the [B]Kalash show stronger affinity to the ancient Scandinavians[/B] than to West Asians. Also, the models more or less reflect the TreeMix analysis above.
Thus, the answer to my question is a resounding yes; there were indeed strong genetic ties between Scandinavia and South Asia during the Bronze Age."
Kalash and the "pheomelanin complex"
https://postimg.cc/mP1gWb8P
https://postimg.cc/QBsGYsPG
Tacitus would have called this without doubt: rutilae comae!
ReplyDeleteChimp.REF Iran_GanjDareh_N Russia_Ust_Ishim.DG Morocco_Iberomaurusian -0.0405 -8.824 53473 57992 928779
result: Chimp.REF Iran_GanjDareh_N ZlatyKun Morocco_Iberomaurusian -0.0369 -7.493 56713 61053 976374
result: Chimp.REF Georgia_Kotias.SG Russia_Ust_Ishim.DG Morocco_Iberomaurusian -0.0365 -6.580 57309 61651 981492
result: Chimp.REF Georgia_Kotias.SG ZlatyKun Morocco_Iberomaurusian -0.0296 -5.164 60795 64502 1033279
IranN and CHG both share more alleles with something basal like ZlatyKun and UstIshim than Taforalt. Z-Scores negative and significant.
@Rob
ReplyDelete"Thus lack of hg E in current Iran N samples is not an issue because (i) they might have acquired north African admixture from Natufians rather than directly from north Africa, and (ii) it is only a level of 20% ('only', but still rather significant). Also, this is marginally higher than in CHG, and much higher than AHG (which are ~ 0%)"
In truth, although skeptical, I don't have a strong opinion on whether Iran_N has Taforalt-related ancestry or not. However, if so I am assuming they have 20% Taforalt-like ancestry instead of 20% of the ANA component?
And that would mean the rest of Iran_N ancestry is paleo-European/West Eurasian and the ANE/ENA combo?
@Cy Tolliver
Yeah for Natufians, Anatolia_N it would make sense. I think one can really use Zlaty Kun as well as AHG/WHG etc in formal statistics to sort of check against the results of any qpGraph output.
https://populationgenomics.blog/2019/05/23/the-big-picture-for-west-asia-and-specifically-anatolia-after-pinarbasi/
ReplyDeleteChad has done some qpgraphs with Natufian, PPN, anatolian, IranN.. Nowhere in his graphs is an IBM or Natufian input seen in IranN.
This comment has been removed by the author.
ReplyDelete
ReplyDelete@ Vasishta
''IranN and CHG both share more alleles with something basal like ZlatyKun and UstIshim than Taforalt. Z-Scores negative and significant.''
those stats do not exclude some form of north African admixture in Iran_N.
For ex
result: Chimp.REF Russia_Sarmatian.SG Russia_Srubnaya Turkmenistan_Gonur_BA_1 -0.0266 -12.051 41809 44098 781388
Sarmatians ''share more alleles with Srubnaya'', but they still have some form of BMAC -related admixture
As a side note - many of the proposals which you boast will be disproven, if not already so. For ex you claim that only a migration from Iran can explain proto-Anatolians, depiste the fact that you very well know about Kum-4, and that Iran consistenly experienced in-migrations post Neolithic. And that's just the tip of the iceberg
I think you'd benefit from building up a more adequate knowlegde base and more honesty instead of an all-consuming desire to 'prove' that homo sapiens come from India
@ Vasistha
ReplyDelete''IranN and CHG both share more alleles with something basal like ZlatyKun and UstIshim than Taforalt. Z-Scores negative and significant.''
Good for chad. Btw some crucial aDNA has been published since 2019, if you hadnt noticed
''result: Chimp.REF Georgia_Kotias.SG Russia_Ust_Ishim.DG Morocco_Iberomaurusian -0.0365 -6.580 57309 61651 981492
result: Chimp.REF Georgia_Kotias.SG ZlatyKun Morocco_Iberomaurusian -0.0296 -5.164 60795 64502 1033279
IranN and CHG both share more alleles with something basal like ZlatyKun and UstIshim than Taforalt. Z-Scores negative and significant.''
So what ?
For ex
result: Chimp.REF Russia_Sarmatian.SG Turkmenistan_Gonur_BA_1 Russia_Srubnaya 0.0266 12.051 44098 41809 781388
Sarmatians ''share more alleles with Srubnaja' but that doesnt mean they lack some form of BMAC-related admixture
@ Gamerz
''In truth, although skeptical, I don't have a strong opinion on whether Iran_N has Taforalt-related ancestry or not. However, if so I am assuming they have 20% Taforalt-like ancestry instead of 20% of the ANA component?''
Neither did I, as my interest in North Africa had until recently been passing. However, it keeps coming up.
Again, based on archaeology, it makes sense, because Epipaleolithic West Asians largely come from near Sinai. This impacted as far as Iran, because the Zarzian represents a population replacement (but don;t tell Vasistha, as he is under the impression that homo sapiens come from india )
@Vasistha
ReplyDeleteBut isnt's UstIshim some sort of early IUP/ENA? Or would that be Bacho Kiro/Tianyuan?
Btw, if you have time how do these come out?
(Chimp,Iran_N/CHG;Anatolia_HG,Taforalt) or/and (Chimp,Iran_N/CHG;WHG,Taforalt)
"Chad has done some qpgraphs with Natufian, PPN, anatolian, IranN.. Nowhere in his graphs is an IBM or Natufian input seen in IranN."
He had posted here last April claiming African/Taforalt input in Anatolia_HG, not sure if he still thinks that's the case.
The source of Late Paleo West Asians
ReplyDelete@Rob
ReplyDelete"Sarmatians ''share more alleles with Srubnaja' but that doesnt mean they lack some form of BMAC-related admixture"
What a strawman. you claimed there is NO (0, zilch, nada) basal ancestry in IranN and that all of this basal looking ancestry comes from ANA. That is demonstrably false.
"For ex you claim that only a migration from Iran can explain proto-Anatolians, depiste the fact that you very well know about Kum-4, and that Iran consistenly experienced in-migrations post Neolithic."
Kum4 the extremely low coverage kumtepe sample with 12000 SNPs? i dont care about that, in any case, lay out what you mean rather than useless remarks.
"I think you'd benefit from building up a more adequate knowlegde base and more honesty instead of an all-consuming desire to 'prove' that homo sapiens come from India"
The fk. I believe in OoA. That is why in all my graphs you will see an OoA node. You're embarrassing yourself.
@ vasistha
ReplyDelete''What a strawman. you claimed there is NO (0, zilch, nada) basal ancestry in IranN and that all of this basal looking ancestry comes from ANA. That is demonstrably false.''
Nope, i said 'basal Eurasian' is an outmoded statistical construct which is no longer required. Like I think WHG is an outmoded concept
but what I specifically said was - I don’t think there is a singular population of “basal eurasians”. It probably represents ancestry streams shared by certain Near Eastern populations during the late paleolithic, with sometimes deeply divergent origins..form Europe to north Africa
It's written right here on this thread, so read carefully instead of embarrasing yourself
'Kum4 the extremely low coverage kumtepe sample with 12000 SNPs? i dont care about that, in any case, lay out what you mean rather than useless remarks.'
You keep claiming that proto-Anatolians come from Iran (despite all the evidence pointing to migrations into Iran during this time). On the other hand, there is strong evidence of migration from Europe into Anatolia precisely when required, and it;ll be unabiguously European, so you won't be able to BS a ghost Iran or Andaman origin for them
It's just another example of how out of your depth you are. Why ? because you lack knowledge and are consumed by bias. The solution - smile and learn
@gamerj_z
ReplyDelete"Btw, if you have time how do these come out?
(Chimp,Iran_N/CHG;Anatolia_HG,Taforalt) or/and (Chimp,Iran_N/CHG;WHG,Taforalt)"
Ill run them now but already im predicting significantly -ve Z Scores. Because Iran and Pinarbasi both descend chiefly from West Eurasians, same as WHG.
@gamerj_z
ReplyDeleteIranN &CHG shares more alleles with Pinarbasi, england_meso & BachoKiro than Taforalt. (possibly more with BK than ZK/UstIshim too given the larger -ve Z scores)
result: Chimp.REF Iran_GanjDareh_N Turkey_Epipaleolithic Morocco_Iberomaurusian -0.1120 -27.140 40429 50623 777116
result: Chimp.REF Iran_GanjDareh_N England_Mesolithic Morocco_Iberomaurusian -0.1087 -27.291 38695 48134 737666
result: Chimp.REF Iran_GanjDareh_N BK-1653_merge Morocco_Iberomaurusian -0.0610 -11.685 27194 30727 526712
result: Chimp.REF Georgia_Kotias.SG Turkey_Epipaleolithic Morocco_Iberomaurusian -0.1234 -24.573 41383 53034 800088
result: Chimp.REF Georgia_Kotias.SG England_Mesolithic Morocco_Iberomaurusian -0.1198 -24.292 38994 49613 747168
result: Chimp.REF Georgia_Kotias.SG BK-1653_merge Morocco_Iberomaurusian -0.0753 -12.018 27895 32437 545106
@ Rob
ReplyDeleteI don't care what you think about the term 'basal Eurasian. Maybe it triggers you, I don't care. It is just a node in the qpgraph for me and it is required to explain IranN and CHG. This node is not Natufian or Taforalt nor is it close to any of these 2. End of.
"You keep claiming that proto-Anatolians come from Iran (despite all the evidence pointing to migrations into Iran during this time). On the other hand, there is strong evidence of migration from Europe into Anatolia precisely when required, and it'll be unambiguously European, so you won't be able to BS a ghost Iran or Andaman origin for them"
There's no need for any ghost. On a simple PCA, one can clearly shift Anatolian populations shift 50% from BarcinN towards Iranian/Armenian populations starting with chalcolithic. If you had any honesty in you, you would acknowledge it. Literally, every paper acknowledges this shift.
Time transect of Anatolia.
Boncuklu_N 8000bce (not great model, suggest better)
Pinarbasi 87%
IranN 13%
p-val: 0.011 https://pastebin.com/peKLqXLf
BarcinN 6300BCE (not great model, suggest better)
Boncuklu_N 76%
IranN 3%
PPN 21%
p-val 0.045 https://pastebin.com/n0P4Mvvd
TellKurdu_EC 5600bce
Turkey_N 54%
PPN 27%
Iran_HajjiFiruz_C 19%
p-value 0.31 https://pastebin.com/1V8MK3r5
Barcin_C 3750bce
Barcin_N 40%
KuraAraxes 60%
p-value 0.06 https://pastebin.com/sUAAJDZe
ArslanTepe_LateC 3750-3100 bce
Turkey_TellKurdu_EC 50%
KuraAraxes 50%
p-value: 0.058
https://pastebin.com/cLDUABb2
ArslanTepe_EBA 2700 bce
Turkey_Arslantepe_LateC 89%
KuraAraxes 11%
p-val: 0.049 https://pastebin.com/Xusqgw7J
Turkey_EBA (Isparta) 2500bce
Turkey_TellKurdu_EC 52%
Armenia_EBA_KuraAraxes 48%
p-val: 0.053 https://pastebin.com/WGyE53ig
This ancestry remains constant till the old Hittite samples MA2200, MA2203. Proto Anatolian already formed between 4000-3000bce. I see no need for any European ancestry, only ancestry entering Anatolia is from its east explains the language.
Now show me your unambiguous European ancestry in Anatolia please lol. All you do is talk, no proof.
Is the Siberian and green eye thing real or just another place where the correlation is indirect or not causal.
ReplyDeleteI don't have a clue what the green eye/Siberian association means and how it came about exactly.
ReplyDeleteBut I wouldn't be surprised if it was just something like both green eyes and Siberian ancestry peaking in northeastern Estonia.
The green eyes association could be just a product of the lower frequency of the derived allele for SLC45a2, as Siberian populations have a lower frequency of the derived allele and I have seen a paper that described a decision tree in which the presence of the allele derived for the SLC45a2 determined whether the eye was blue or green.
ReplyDelete@Rob
ReplyDelete"Neither did I, as my interest in North Africa had until recently been passing. However, it keeps coming up.
Again, based on archaeology, it makes sense, because Epipaleolithic West Asians largely come from near Sinai. This impacted as far as Iran, because the Zarzian represents a population replacement"
But based on archaeology it doesn't seem the Zarzian is coming from the south though right? Rather from the west IIRC. That doesn't preclude admixture, in any case but I think you won't find substantial Taforalt-like ancestry in say Kebaran samples if they are ever found. Btw, what's Taforalt made up of then? Do you agree with the model that it is a mix of largely Anatolia_HG/Dudzuana-like people and "ANA" (whatever ANA is)?
@Vasistha
ReplyDeleteThanks for the stats. Well, I was trying to think of a way to see whether Iran_N is more shifted to Taforalt than Anatolia_HG or WHG, I am not sure what would work. Would something like (Chimp,CHG;Iran_N,Taforalt) work or would any basal admixture (and the Western part of Taforalt) mess this up?
Alternatively, maybe running a qpAdm on Iran_N could point whether they have Taforalt-like admix or not.
Actually if you feel like it, what would Natufians look like in the stats you posted in place of Iran_N for example?
@ Vasistha
ReplyDelete“ On a simple PCA, one can clearly shift Anatolian populations shift 50% from BarcinN towards Iranian/Armenian populations starting with chalcolithic”
Understand this , bozo
- we’re not talking about Arslantepe; we are saying that there is / will be evidence of European ancestry in western -central Anatolia
- the so-called Iranian ancestry in Chalcolithic Turkey is from the Caucasus and Mesopotamia, not on out of Iran or India migration. Your bias compels you to muddy the waters
“ I don't care what you think about the term 'basal Eurasian. Maybe it triggers you, I don't care.
Verbal diarrhoea of a brat claiming he’s the king of admixtools. You’re just blindly manicuring data of samples and contexts you have no idea about .
It’s only required in your models because they’re all about ghosts and make-believe.
Also, wasn't there an Italian study that pretty much stated increasing steppe ancestry wasn't associated with phenotypical change in Italy?
ReplyDeleteThis Estonian paper is not the only paper debunking the whole steppe = blonde hair thing.
@rob
ReplyDelete"we’re not talking about Arslantepe; we are saying that there is / will be evidence of European ancestry in western -central Anatolia"
Go to the map and check where Barcin is. We already have chalcolithic & bronze age samples from central Anatolia. Show your proof rather than insulting me like a child.
We also have central anatolian samples from the hittite contexts, they are very similar to other chalc and EBA samples.
Your theories are dead on arrival.
@Rob
ReplyDeleteStop acting all high and mighty, you keep blabbering without giving proof or running a single model. Thats how arrogant you are.
Here's western Anatolia for you, your precious Kumtepe Kum6.
Turkey_Kumtepe_N.SG (3600bce)
Turkey_N 60%
Armenia_EBA_KuraAraxes 40%
p-val: 0.488
right pops:
Mbuti.DG
ONG.SG
EHG
CHG
England_Mesolithic
Morocco_Iberomaurusian
WSHG
Tarim_EMBA1
Mongolia_North_N
Turkey_Epipaleolithic
Iran_GanjDareh_N
PPN
Russia_Steppe_Eneolithic
Vasistha - actually Dave and I have patiently & repeatedly informed you that it’s established, not to prove you wrong but to help you But you insist on making a fool of yourself, and are doing so brilliantly. Doubly so with how arrogant you are.
ReplyDeleteSo here’s a shovel- keep digging
@Rob
ReplyDeleteI don't care about your ideologies and fantasies. I deal with facts on the ground. Numbers, models - not your banal words.
The eastern provenance of Anatolian languages has been proposed by many, and the genetic data bears that out. That is all there is to it.
Kura Araxes seems like a very very good contender, given 40-55% of it in Anatolia stating 4000bce.
http://www.spekali.tsu.ge/index.php/en/article/viewArticle/2/16
The genetic data agrees with Nikoloz Silagadze's theory of KA origin of Anatolian languages. Archaeology confirms the KA genetic input into Anatolia.
I rest my case, wont respond to you further on this topic.
This comment has been removed by the author.
ReplyDeleteWell, there's paternal steppe ancestry in Kura-Araxes anyway.
ReplyDeleteSo that's obviously its Indo-European link.
https://eurogenes.blogspot.com/2018/12/r-v1636-eneolithic-steppe-kura-araxes.html
Eneolithic Steppe is Indo-European now?
ReplyDeleteThere's V1636 in Single Grave, and Kura-Araxes is about the same age.
ReplyDeleteSo whether all of Eneolithic steppe was Indo-European or not, the V1636 in Kura-Araxes looks to be part of the Indo-European expansions.
@Rob
ReplyDelete"And note, modelling it with Kura-Araxes, a culture which dates to 3000-2200 BC, is highly ahistoric."
As usual, you lie and deceive. The standard dating of Kura Araxes so far has been 3500bce onwards. The Kura araxes samples themselves (arm001 & arm002) are dated between 3600-3100bce.
Plus, new KA site pushes back the earliest date to last quarter of 5th mill bce.
On the Genesis of the Kura-Araxes phenomenon: New evidence from Nakhchivan
"THE EARLIEST KURA-ARAXES ARTEFACTS KNOWN TO DATE: THE EVIDENCE FROM OVÇULAR TEPESI (CA 4300-4100 BC) "
So as usual, you shoot from your butt to defend your fantasies without knowing the facts.
I wouldn't be so hasty in assigning V1636 to an Indo-European expansion, considering that the oldest confirmed Indo-European expansion into South Caucasus is linked with Martkopi-Trialeti. V1636 is roughly contemporary with the aforementioned cultures, but the Steppe ancestry in I1635 is highly diluted, and in turn could be a leftover from the Steppe-rich Areni-like folk, along with the paternal marker.
ReplyDeleteOn top of that, I don't see any signs of Ukrainian forager ancestry in I1635, whose Steppe appears to be exclusively Progress-like, so that also opens up another Pandora's box.
ReplyDeleteKA is a very different culture to CWC or Usatovo., & V3616 isn’t a particularly relevant marker. In fact, the eastern /: Volga steppe groups might not have been IE
Whatever steppe ancestry there is in KA or Areni C appears to have become culturally assimilated by locals
The situation is different in the post-KA phase, when Katacomb groups moved south and established themselves in the Caucasus. They’re arguably early Armenians, although that still needs clarification
@Rob
ReplyDelete"What I said above was Kum-4, the much younger sample, coincident with creation of Troy, and it represents a small glimpse of what is to come."
It is irrelevant, the much later Hittite period samples show no extra ancestry than what I have already described. Isparta EBA, central Anatolia (2400bce) also lacks your ancestry.
Stop making mountains out of a molehill from some single 12000snp sample sheesh. Wonder why it's even there on G25 lol. Maybe Davidski is playing with people like you as his audience.
@Davidski
ReplyDelete"There's V1636 in Single Grave, and Kura-Araxes is about the same age.
So whether all of Eneolithic steppe was Indo-European or not, the V1636 in Kura-Araxes looks to be part of the Indo-European expansions."
The V1636 sample is late KA, near the end of KA culture.
Anyway, the KA autosomal ancestry doesn't quite need Progress ancestry.
Vahaduo results
https://imgur.com/a/7e8mWYc
There are new regional samples from modern Republic Armenia ( Syunik province ).
ReplyDeleteTarget: Armenian_Syunik
Distance: 0.0287% / 0.02866348
46.8 TUR_Tepecik_Ciftlik_N
19.7 IRN_Ganj_Dareh_N
17.8 GEO_CHG
9.2 Yamnaya_RUS_Samara
6.5 Levant_PPNB
Target: Armenian
Distance: 0.0288% / 0.02876767
46.4 TUR_Tepecik_Ciftlik_N
19.5 IRN_Ganj_Dareh_N
19.0 GEO_CHG
10.1 Levant_PPNB
5.0 Yamnaya_RUS_Samara
When South Caucasian Neolithic is added the models improve. The Steppe ancestry increase. As I stated here many times the cryptic WHG-ish influences both in CHG and Anatolia affects most strongly the Armenian results because Armenians do not have true ancestry from this pops but rather something basal like Çayonu and their derivatives like Caucasian Neolithic.
Target: Armenian_Syunik
Distance: 0.0212% / 0.02116688
69.3 AZE_Caucasus_lowlands_LN
12.8 Yamnaya_RUS_Samara
10.2 TUR_Tepecik_Ciftlik_N
6.9 IRN_Ganj_Dareh_N
0.8 Levant_PPNB
Target: Armenian
Distance: 0.0197% / 0.01974371
74.9 AZE_Caucasus_lowlands_LN
8.7 Yamnaya_RUS_Samara
6.8 TUR_Tepecik_Ciftlik_N
5.5 IRN_Ganj_Dareh_N
3.9 Levant_PPNB
0.2 Han
@Davidski" I don't have a clue what the green eye/Siberian association means and how it came about exactly.
ReplyDeleteBut I wouldn't be surprised if it was just something like both green eyes and Siberian ancestry peaking in northeastern Estonia."
First of all the recent paper gives a positive correlation between green eyes (that's pheomelanin to the max) and Siberia/ENA and a negative one with EEF.
A paper from Peter Frost, The Puzzle of European Hair, Eye, and Skin Color (2014) is illuminating in this respect. In fact he states:
1. Sexual selection plays a crucial part, and MC1R plays an essential part in it, because it gives more color in the palet, green eyes, amber eyes, red hair, warm blond! Tempting.... ;)
2. Especially area's like Siberia (but higher latitude in general) play a crucial part in it.
So I tend to think that it was ANE and so indirect the Steppe component who brought it into the Celto-Germanic range. The Irish and Scottish red heads and the North Sea Germanic warm blonds....
https://www.academia.edu/7110502/The_Puzzle_of_European_Hair_Eye_and_Skin_Color
I keep seeing posts regarding jat ancestry models showing 40% steppe.
ReplyDeleteHaplogroup L (36.8%)
Haplogroup R (28.5%)
Haplogroup Q (15.6%)
Haplogroup J (9.6%)
Haplogroups E, G, H, I, T (9.5%)
Do Jats show autosomal East Asian ancestry? Any idea which R sub-clade they are? Any Andronovo connection? Or is it mostly related to Scythian-Huns?
Traditionally been pastrolists who took to farming and only in last few centuries became politically active.
@vAsiSTha
ReplyDeleteI said paternal steppe ancestry.
And there's an earlier V1636 in Anatolia.
It makes no difference to me whether these samples with paternal steppe ancestry had steppe autosomal ancestry or not.
@davidski
ReplyDeleteWell that's quite shortsighted lol. Y dna can travel randomly too
@DragonHermit, yes, there was that finding that there was no strong differentiation associated in Italy with the introduction of steppe ancestry.
ReplyDelete"Debunked"? Well, from the perspective of ancient dna the proposal that early steppe groups were *very* highly blonde and this was reduced down in frequency to present day levels by admixture (then there was no or little selection afterwards), that idea was never really "bunked" in the first place as soon as they were looking at pigmentation linked variants.
E.g. https://www.cell.com/current-biology/pdf/S0960-9822(19)30424-5.pdf - The Arrival of Siberian Ancestry Connecting the Eastern Baltic to Uralic Speakers further East - Notably, we infer a high proportion (40%–60%) of dark skin pigmentation in the hunter-gatherers and CWC farmers (Data S2). We infer dark skin and blue eyes for two individuals, similarly to another European Mesolithic individual [39]. However, from BA onward, we infer pale or intermediate skin pigmentation for all individuals and an increase in the proportion of blue eyes and lighter shades of hair (Data S2). This is in line with previous suggestions that light skin pigmentation alleles reached high frequencies in Europe only recently [40].
(When this paper was published, a commentor noted here that blonde hair is more frequent in Iron Age Estonian samples with Siberian ancestry - https://imgur.com/a/El3i6Bf. Which indicates that the presence of Siberian admixture in the population that was present then (either the descendent of the Bronze Age one or new), was not enough to offset a greater selective effect in time from BA->IA groups.)
Similar finding in Fatyanovo - https://www.science.org/doi/10.1126/sciadv.abd6535 (see Table 2).
I would make the distinction that this idea (being predominantly blonde and blue eyed, reduced by admixture) is not the same, as whether the steppe groups were on average more likely to express blondeness than the average contemporary EEF group. This is an idea that is still fairly plausible based on adna - it could've been the case that most EEF groups had darker hair on average.
However, I think we only really have the evidence of increase in these variants in later mixed steppe-EEF groups, not the initial steppe migrants, and like I indicate above, I think it's possible there could've been some degree of selection by picking and choosing women with more light brown/blonde-ish hair from EEF groups (which might be one thing that could have created this association that Pagani's group seem to find here). If there were marriages between CWC and previous European farmers to "keep the peace" that went the way of predominantly male CWC and female EEF, with the luxury to choose they may well have been choosing women from the EEF group with rarer hair colour variations. (This idea can definitely be tested if we start to find more evidence of female EEF outliers in CWC sites, or their first generation sons and daughters). But anyway the bulk of change must have been selective forces over time.
@Tigran, my take is that it's possible that Siberian groups could've introduced East Asian variants that push eye colour from dark brown to honey brown, and these may (with the influence of selection) be causal to green eye colour in Estonians today. This is in light of Siberian ancestry also being enriched for dark eye colour associated variants. But this would need some more research and description to check if its true.
ReplyDeleteWe definitely shouldn't be take these associations as indicating that there was a "green-eyed Siberian" or "blonde-haired EEF" population in the past. They're just indicating that the linked variants seem to be disproportionately enriched for EEF/WHG/Steppe/Siberian ancestry around them and there could be various reasons for this. Not that they didn't need selection to get to their present day frequency, nor to predict the phenotype in the ancestral populations.
The ancient dna does indicate that if the mixed Siberian/Steppe population that dispersed to Estonia did form in Russia around 1500-1000 BCE prior to dispersing to Europe in the Iron Age, there might have been less difference in eye, hair and skin colour between the Siberian and Steppe groups that met in Russia before dispersing to Estonia / Finland, compared to the groups in Estonia / Finland today. May have been more like predominantly dark blue/grey eyed and dark haired Steppe people meeting with black haired and brown eyed Siberians. Though the differences would already have likely begun to emerge by this time already.
@ancestralwhispers.org Is your avatar a reconstruction of Steppe Maykop?
ReplyDelete@ Matt from Frost:
ReplyDelete"For others still, this color diversity arose through random factors: genetic drift, founder effects, relaxation of natural selection, etc. But these factors could not have produced such a wide variety of hair and eye hues in the 35,000 years that modern humans have inhabited Europe. The hair-color gene (MC1R) has at least 7 alleles that exist only in Europe and the same is probably true for the eye-color gene (OCA2). If we take the hypothesis of a relaxation of selection, nearly a million years would be needed to accumulate this amount of diversity. Moreover, it is odd that the same sort of diversification has evolved at two different genes whose only point in common is to color a facial feature."
@Matt
ReplyDeleteInteresting. How would that lead to green eyes though? I am sure West Eurasians have their own SNPs that lead to light brown eyes,
''"THE EARLIEST KURA-ARAXES ARTEFACTS KNOWN TO DATE: THE EVIDENCE FROM OVÇULAR TEPESI (CA 4300-4100 BC) "''
ReplyDeleteActually, Marro's dating is disputed and lone voice. 4300 bc is still Chaff Faced Ware culture, not Kura-Araxes
Obviously, the Calcuttan Archie isn't cognizant of all facts, issues and details, just randomly googles & quotes without digesting
@weure
ReplyDeleteFrost has a psychosexual bias in his work, he is biased by fetishism, his dumpster fire blue-eye brown-eye paper concerning Czech university students is an example of this, sexual selection is possible but I would take any Frost work with a grain of salt.
I noticed some discussion of haplogroup R1b1a2-V1636 in this thread.
ReplyDeleteThe public Y-DNA tree at 23mofang, a direct-to-consumer genetic testing company based in Chengdu, Sichuan, currently includes a total of 928 members of haplogroup R1b-M343 (TMRCA 18650 years). This amounts to 0.415% of the entire database.
The most common subclade among members of R1b-M343 on 23mofang is Western European R1b1a1b1a-L51 (TMRCA 5640 years) with a total of 252 members; however, a majority of these individuals appear to be foreigners who somehow have purchased a test from 23mofang or otherwise patrilineal descendants of Western Europeans who happen to live in the PRC (e.g. descendants of Englishmen in Hong Kong or Portuguese in Macau).
Excluding R-L51 leaves a total of 676 members of R1b-M343, amounting to approximately 0.3% of the entire database.
This "non-Western European" R1b segment of the Chinese population may be further analyzed as follows:
175/676 = 25.89% R1b2-PH155 (TMRCA 7400 years)
153/676 = 22.63% R1b1a1a1-M478 (TMRCA 7790 years)
116/676 = 17.16% R1b1a1b1b-Z2103 (TMRCA 4910 years)
97/676 = 14.35% R1b1a1b1-L23* (TMRCA 5720 years)
60/676 = 8.88% R1b1a2-V1636 (TMRCA 6720 years)
35/676 = 5.18% R1b1a1b-M269* (TMRCA 6300 years)
18/676 = 2.66% R1b1-L754* (TMRCA 17230 years)
8/676 = 1.18% R1b-M343* (TMRCA 18650 years)
6/676 = 0.89% R1b1b-PF6279/V88 (TMRCA 11960 years)
4/676 = 0.59% R1b1a1b2a-GG480 (TMRCA 4600 years)
2/676 = 0.30% R1b1a1-P297* (TMRCA 13950 years)
2/676 = 0.30% R1b1a1b2-PF7558* (TMRCA 5400 years)
The subclade composition of the R1b-M343(xL51) segment of the Chinese population does not suggest a likelihood of recent origin from Western Europe. The surnames and geographic distribution of these individuals within China tend to suggest proximally Mongol, Manchu, Turkic (e.g. Uyghur), or just generally Muslim (e.g. Hui) backgrounds for many of these individuals.
The prevalence of R1b2-PH155 and R1b1a1a1-M478 among eastern members of Y-DNA haplogroup R1b has been noted previously, but I was a bit surprised to see that the proportion of Chinese R1b that belongs to R1b1a2-V1636 also may be greater than the proportion of European R1b that belongs to R1b1a2-V1636.
@Andrzejewski
ReplyDelete"Is your avatar a reconstruction of Steppe Maykop?"
No, Dolmen culture.
@Ebizur
ReplyDelete"but I was a bit surprised to see that the proportion of Chinese R1b that belongs to R1b1a2-V1636 also may be greater than the proportion of European R1b that belongs to R1b1a2-V1636."
Well, I'd expect that rarer branches have higher proportion compared to other R1b branches in China than in Europe, because in Europe the dominant branches had huge founder effects and therefore have very high modern frequency, while in China there were no such huge founder effects of R1b branches, so the overall frequency (the base of comparison for a proportion) is much lower. So it is not that V1636 have anything special going on in China, there are just no big branches to overshadow it within R1b.
@Aut-Right I'm the last one who takes everything from Frost without salt....on the contrary.
ReplyDeleteBut in the case he has imo a point, sexual selection must have been a force that contributed to the diversity in hair and eye color in Europe. The key is: 'If we take the hypothesis of a relaxation of selection, nearly a million years would be needed to accumulate this amount of diversity' is imo plain logic. And feel free to debunk it.
By the way, the total number of entries on the public version of the 23mofang phylogenetic tree (≈222,200) is comparable to the total number of entries on the public version of Y-DNA Haplotree at FTDNA (217,509).
ReplyDeleteThe Y-DNA Haplotree at FTDNA currently has a total of 46 members tabulated under haplogroup R-V1636, whereas 23mofang currently has a total of 60 members tabulated under R-V1636, so it appears that R-V1636 should be at least as common in China as it is in Western Eurasia.
The geographic breakdown of the members of R-V1636 on the Y-DNA Haplotree at FTDNA is as follows:
Kuwait x4, Italy x2, Lebanon x2, Belarus x2, Puerto Rico x2, Armenia x1, Russian Federation x1, Canada x1, Germany x1, Turkey x1, Saudi Arabia x1, Qatar x1, Bulgaria x1, Dominican Republic x1, Iraq x1, Unknown Origin x24
The members of R-V1636 at 23mofang are as follows:
R1b-M343 > R-L754 > R-L389 (TMRCA 15710 years) > R-V1636 (TMRCA 6720 years) 52 members (Buhe 布和 from Xilingol League, Inner Mongolia x1, Xie from Datong, Shanxi x1, Jiang from Wuxi, Jiangsu x1, Li 李 from Beijing x7, Xu from Datong, Shanxi x1, Wang from Yancheng, Jiangsu x1, Lian from Linfen, Shanxi x1, Ji 吉 from Lyuliang, Shanxi x1, Li from Chifeng, Inner Mongolia x1, Liu from Baoding, Hebei x1, Zhang from Bazhong, Sichuan x2, Zhang from Xinzhou, Shanxi x1, Liu from Cangzhou, Hebei x2, Wang from Baoding, Hebei x1, Liu from Zhumadian, Henan x1, Kong from Jining, Shandong x2, Gu from Wuxi, Jiangsu x1, Zhang from Beijing x2, Wei from Shijiazhuang, Hebei x1, Wang from Zhongwei, Ningxia x1, Cao from Nanjing, Jiangsu x1, Su from Yuncheng, Shanxi x1, Ren from Xinxiang, Henan x1, Wang from Tieling, Liaoning x1, Shen from Ulanqab, Inner Mongolia x1, Tang from Shanghai x1, Yang from Xuzhou, Jiangsu x1, Ma from Changchun, Jilin x1, Xing from Tianshui, Gansu x1, Xin from Hohhot, Inner Mongolia x1, Xu from Baoding, Hebei x1, Chang from Xilingol League, Inner Mongolia x1, Zheng from Jinzhong, Shanxi x1, Chen from Zhuzhou, Hunan x1, Hao from Xinxiang, Henan x1, Tong from Shanghai x1, Ma from Haidong, Qinghai x1, Han from Tianjin x1, Wang from Suzhou, Jiangsu x1, Zhang from Xi'an, Shaanxi x1, Ji 冀 from Beijing x1, Li from Shenyang, Liaoning x1)
R1b-M343 > R-L754 > R-L389 > R-V1636 > R-V1274 (TMRCA 6720 years) 1 member (Ma 马 from Luohe, Henan x1)
R1b-M343 > R-L754 > R-L389 > R-V1636 > R-V1274 > R-MF225042 1 member (Li 李 Hui from Chifeng, Inner Mongolia x1)
R1b-M343 > R-L754 > R-L389 > R-V1636 > R-V1274 > R-BY15352 > R-MF239526 (TMRCA 2250 years) 2 members (Guo 郭 Han Chinese from Anshan, Liaoning x1, Zhang 张 Han Chinese from Qingyang, Gansu x1)
R1b-M343 > R-L754 > R-L389 > R-V1636 > R-Y106006 > R-Y98084 (TMRCA 3020 years) > R-MFV1260 1 member (Ma 马 Hui from Haidong, Qinghai x1)
R1b-M343 > R-L754 > R-L389 > R-V1636 > R-Y106006 > R-Y135345 > R-Y183240 (TMRCA 3530 years) 1 member (Ma 马 Hui from Linxia Hui AP, Gansu x1)
R1b-M343 > R-L754 > R-L389 > R-V1636 > R-Y106006 > R-Y135345 > R-Y183240 > R-MF235797 (TMRCA 3530 years) 2 members (Su 苏 Han Chinese from Yuncheng, Shanxi x1, Zhang 张 "Other" from Beijing x1)
60 members R1b1a2-V1636 total (TMRCA 6720 years)
The members of R-V1636 (formed 15600 [95% CI 17600 <-> 13800] ybp, TMRCA 6600 [95% CI 7700 <-> 5700] ybp) currently tabulated on the YFull tree are as follows:
ReplyDeleteChina x8 ("Dungan" i.e. Hui from Qinghai x2, "Dungan" from Inner Mongolia x1, "Mandarin Chinese" from Liaoning x1, "Mandarin Chinese" from Beijing x1, "Jinyu Chinese" from Shanxi x1, Gansu x1, "Tajik" from Xinjiang x1; however, I am a bit suspicious that this "Tajik" individual may in fact be a "Lowland Tajik" from Dushanbe, Tajikistan [cf. Min-Sheng Peng et al. 2017, "Mitochondrial genomes uncover the maternal history of the Pamir populations"] who may have been mislabeled by YFull)
Kuwait x5 (Four of these Kuwaitis share a common ancestor with one of the Qatari individuals 650 [95% CI 1000 <-> 325] ybp.)
Turkey x3 (One of these is indicated to be a speaker of "North Mesopotamian Arabic" from Mardin.)
Qatar x2
Belarus (Gomel Region) x1
Bulgaria (Blagoevgrad) x1
Finland (Southern Finland Province) x1
Iraq (Al Anbar) x1
Israel (HaMerkaz) x1
Italy (Salerno) x1
Puerto Rico x1
Russia (Tomsk Tatar) x1
Saudi Arabia ("Arabic") x1
Switzerland (Valais) x1
Ebizur
ReplyDeleteIn ArmDNA project there are three cases of V1636. But only one was tested for that SNP. Other two cases are predicted by STR.
https://www.familytreedna.com/public/ArmeniaDNAProject?iframe=yresults
Aram wrote,
ReplyDelete"In ArmDNA project there are three cases of V1636. But only one was tested for that SNP. Other two cases are predicted by STR."
The SNP-tested individual (a certain Mr. Vardapetian from Armenia) may be the R-V1636 individual from Armenia on the Y-DNA Haplotree at FTDNA.
The data from the Armenia DNA Project at FTDNA suggest that perhaps 0.21% (3/1436) of Armenians may belong to R-V1636.
The data from 23mofang suggest that perhaps 0.027% of Chinese may belong to R-V1636.
These figures suggest that R-V1636 in Armenia may be nearly eight times as common as R-V1636 in China. However, with only three members of this clade from Armenia (only one of whom has actually been tested for the V1636 SNP), this result should be considered tentative.
@Tigran, its all a continuous phenotype, worth reading through this paper - https://www.nature.com/articles/s41467-018-08147-0.
ReplyDelete@weure, I can't comment on Frost's time estimate as he does not explain it, though I think he's not the most trustworthy source.
He had big problems with the adna showing that strong selection on light coloured eyes might have happened within the last 3000 years, rather than in his preferred Upper Paleolithic period, mostly for ideological reasons. And some big problems with the idea of replacement of European HG populations (https://evoandproud.blogspot.com/2010/10/ich-bin-mittel-ostlander.html / https://evoandproud.blogspot.com/2011/12/were-native-europeans-replaced.html) essentially not on the evidence but because it contradicted his theory of selection for blondeness being selection for female beauty during the Upper Paleolithic. As late as 2018, he appeared to reject essentially rejects a majority genetic replacement of European HG on non-senseical grounds and bizarrely propose that Europeans are descended from Scandinavan or Baltic Mesolithic HG (https://evoandproud.blogspot.com/2018/02/cheddar-man.html).
Anyway, I think few people are saying it is necessarily just relaxed constraint, rather that the variants could've got to their frequency through relaxed constraint, drift and very mild levels of selection that are almost undetectable through tests.
I think Frost's at least wrong that these variants are only present in Europe though, anyway.
@Matt clear that he is selling some humbug about the the mittel ostlander, the HG etc. There are more things which leads to frowns.
ReplyDeleteOn the other hand "Frost's time estimate as he does not explain it" well not en detail that's correct. Nevertheless the principle the plain logic is clear. 'If we take the hypothesis of a relaxation of selection, nearly a million years would be needed to accumulate this amount of diversity'.
And with regard to the last paper I guess in this respect he has some point the Siberian/ENA connection with green eyes. He already pointed at that direction (specific Steppe/ Siberia) without the knowledge of the latest paper of course, It's indeed an example of more diversity, more bright/warm colors and behind this the MC1R mutations, we also recognize in the Celto-Germanic population the Irish/ Scottish red heads and the warm blond North Sea Germanics...
@Matt “ I think Frost's at least wrong that these variants are only present in Europe though, anyway.”
ReplyDeleteThat’s why I claim to be a Corded Ware offspring: I acknowledge that I may also be a descendant of farmers and foragers, likely predominantly via GAC, but to me having some of these non-IE bloodlines is akin to someone with 15% Native American admixture claiming to be an American Indian. Particularly when my aDNA is close to 60% Steppe herder.
Hi. How do you determine Sardinian origin? Regards. bauduhin@gmail.com
ReplyDeleteThe interested should check for V3616, M73 etc in a couple of the Xiongnu & Xianbei individuals from Wang, etc ..
ReplyDeleteThis comment has been removed by the author.
ReplyDeleteThe new ancient DNA from bohemia is good for seeing where blonde hair comes from. Farmer or Kurgan.
ReplyDeleteBecause, they got lots of Funnel Beaker & Early Corded Ware DNA.
Arza tested some key SNPs. TRB & early CWC were both more fair skinned than most Europeans of the time. Including Yamnaya. Early CWC had mostly brown eyes, TRB had mostly light eyes.
We can test more SNPs. But maybe I doubt we'd learn anything. It might be best to wait for new studies.
@Andrzejewski
ReplyDelete"Particularly when my aDNA is close to 60% Steppe herder."
Interesting. How did you determine that? Did you run qpAdm on yourself?
What I want to know is how Andrze extracted ancient dna from himself
ReplyDeleteHe's talking about the Polish average based on qpADM run Davidski did last year.
ReplyDeleteI'm planning on doing a detailed video on the distribution of Kurgan ancestry. Both in the present day and circa 0ad.
It sucks there's no one who is overwhelmingly of Kurgan origin today. Whatever region they'd be from, they would be letting us know in the comment section here.
I used early Corded Ware as the steppe proxy in those European qpAdm tests.
ReplyDeleteEarly Corded Ware is much closer to the steppe ancestry in Poles and probably in almost all Europeans than Yamnaya.
@Davidski
ReplyDeleteDavid do you know if the Copenhagen lab has some unpublished Yamnaya specimens with L51 and M269*/PF7562/P297? I was going over Egfjord et al. (2021) again (the one that had the V1636 Single Grave individuals), and in said paper they have a phylogenetic placement chart of R1a and R1b bearing samples. On the chart, it looks like some Yamnaya samples are contained under the L51 and M269*/P297/PF7562 lines. Additionally, is the Yamnaya Caucasus individual SA6010 (Y-DNA: R1b-preV1636) still Yamnaya? The Reich lab oddly changed his label to “Russia_Caucasus_Eneolithic,” despite his carbon date of 2884-2679 calBCE. Their rational for doing so was that SA6010 is a “relative” of SA6001. These two sample have different mtDNA, autosomal DNA (SA6001 is a Steppe Maykop sample), and contradictory carbon dates. Quite strange.
@Simon
ReplyDeleteYou can double check this with Egfjord et al., but my understanding is that all of the Yamnaya samples in their schematic have already been published.
So my guess is that those Yamnaya samples are placed where they are due to incomplete data.
R1b-PF7562 in Yamna would be interesting. It is quite possible that this haplotype moved to Anatolia via Balkans.
ReplyDeleteBut we still lack any aDNA from this branch.
@Simon
ReplyDeleteYou can keep desperately searching for R1b-L51>L151>P312 samples in the steppes and looking for all kinds of arguments to justify an eastern origin of that lineage. Harvard and Max Planck have been doing that for 10 years and so far they have failed so I don't think you will succeed. Why don't you start looking elsewhere? How about these three British samples (Patterson, 2.021)-
*I3035 (3.750 BC-????)-Fox Holes Cave-Neolithic-England-HapY-R1b-U106>DF96>A7208
*I3019 (3.200 AC-????)-Cheddar, Totty Pot-England-HapY-R1b-M269>P310
*I2611 (2.998 BC-????)-Tyne and Wear-England-HapY-R1b-L21>DF13>Z253>A11001
We already know that the Kurganists, every time a sample of this lineage appears in Europe that complicates or demolishes their steppe theory, try by all means to discredit it. It has happened with the cases in Spain (ATP3, CLL007), Norway (VK531), Bulgaria (Smyadovo), Switzerland (Auvernier, Burgaschisee), Austria, etc. In the case of the English samples this is what the researchers tell us
Regarding I2611-"All of the burials identified above are expected to date from the Early Bronze Age, so the Late Neolithic radiocarbon date of 4370±35 BP (3090–2906 cal BCE; Poz-83500) on sample I2611 is much earlier than expected-The use of a consolidant on the teeth may have resulted in contamination affecting the radiocarbon determination. The sample has therefore been excluded from the analysis"-Ha Ha Ha Ha
Regarding I3019-"Palaeogenetic data from an undated loose tooth from an adult right maxilla (2004.9/419; SB403C2), yielded sample I3019 (male) which has been excluded from analysis due to low coverage"-Ha Ha Ha, check the coverage, many poorer quality samples have been accepted and used systematically for all types of analysis without problems.
Regarding I3035-"A human petrous temporal from the cave was successfully analysed for aDNA, yielding sample I3035 (male), thought to date to c. 4000‒3500 BCE"-Ha Ha Ha, I suppose that these gentlemen will agree to re-date that sample-That sample is U106 (it is true that it seems that the SNPs identified seem much younger but, it is necessary to check it).
When you have to discard so many samples because they do not fit the theory you want to prove you have a big problem.
Andrei's last comment deserves a special place among the hundreds of thousands that have been posted on this blog. I can't hide the fact that I like him.
@Rob
ReplyDelete"What I want to know is how Andrze extracted ancient dna from himself"
A legitimate concern indeed, I heard that drilling and grinding your petrous bone can lead to health problems.
Don't know about Andrzej or you guys, but I just spat into a test tube to get my DNA.
ReplyDelete"What I want to know is how Andrze extracted ancient dna from himself"
ReplyDeletehttps://biosciences.gatech.edu/events/digging-ancient-haplotypes-out-modern-human-genomes
@Davidski
ReplyDelete"This is a somewhat curious finding considering that ancient Anatolians weren't particularly blond haired or blued eyed, and that's probably an understatement."
Idealized Hellenes are portrayed as blue-eyed blond people.
Hittites(nothing is more ancient Anatolian than them) by ancient Egyptians were desribed as white skinned, blue eyed and with reddish hair. Unrelated to Anatolians, but modern dirty blond has reddish color and some of those dirty blondes think that they have red hair.
The big question is what is that Anatolian_N and from where it came and what is the time periaod that it applies to?
The way, how it is worded in paper""The contemporary European genetic makeup formed in the last 8,000 years when local Western Hunter-Gatherers (WHGs) mixed with incoming Anatolian Neolithic farmers" - I would have to guess that it has nothing to do with 8000 years, otherwise, why compare Yamnaya, so I would think, that ancient Anatolian as Anatolian_N here are IE speaking people localized in Northern Anatolia(which was mostly populated by Kartvelians up to modern days) - so Anatolian N localization is around Troy - essentially one of the Hellenic people.
Since this blog rules out that IE originated in Anatolia, it also should rule out, that blue eyes and blonde hair originated in Anatolia, but arrived there and hence the modern Estonian relations to Anatolian_N is via something else, that spread Anatolian_N to Anatolia and to north.
I suppose, that Aryan invaders in India also were blue-eyed, as blue-eyed Indians are rare, but still a thing.
"Germanic and Finnish populations are somewhat richer in ancient Anatolian ancestry than Estonians, and, unlike ancient Anatolians, they're often exceptionally blond haired and blue eyed... The more direct link is between Germanic and Finnish ancestry and blond hair and blue eyes."
There is absolutelly no relation to Germans in "Estonian Germans", because their origin(and dna) is Estonian - they switched to German language, because that was language of ruling class.
Since Finnic people initially did not had blue eyes and blonde hair as defining traits(because if they had, then Saami would be blond with blue eyes), they got those from CWC, who initially were responsible for spreading blue eyes and blonde hair in Europe(blue eyed mediterranian race just does not make any sense).
Blonde Finno-Ugric people with blue eyes in Polar and Uralic regions seems to be mostly later Rus mix - for example, a big share of Izhma Komi looks like Russians, that switched to Komi language(or rather Russians settled among Komi, taking Komi wives).
CWC in Baltic was expanding with mtdna H, that mostly replaced other mtdna, so even if there was later influx of Finnic y-dna N in region - including Finland, mtdna H is still prevailing among other mtdna along with traits of eye and hair color. That and from my school level Biology lessons I know, that looks are mix of genes from both parents of fruit flies and that also applies to humans.
@Genos
ReplyDelete"It sucks there's no one who is overwhelmingly of Kurgan origin today"
Speak for yourself I am 80% steppe 😅
In connection with exotic influences in China, two more clades to keep tabs on are L1a1-M27 > L-Y31961 > L-Y31214 > L-Y169295 and G2a-P15 > G-L1259 > G-L30 > G-CTS574 > G-CTS2488 > G-P303 > G-L140 > G-PF3346 > G-PF3345 > G-CTS342 > G-Z724 > G-CTS1475.
ReplyDeleteYFull currently has two members of L-Y169295 (formed 8000 [95% CI 9300 <-> 6800] ybp, TMRCA 1800 [95% CI 2700 <-> 1200] ybp): YF064122 from Chongqing, China and YF019658 from Henan, China.
23mofang currently has 22 members tabulated directly under the L-M27 root (TMRCA 8860 years) and a total of 52 members tabulated under L-Z20087 (TMRCA 1310 years), a subclade of L-Y169295.
L-M20 (TMRCA 14070 years) > L1-M22 (TMRCA 14070 years) > L1a-M2481 (TMRCA 14070 years) > L1a1-M27 (TMRCA 8860 years) 22 members (Ma from Tianjin x1, Wang from Xuchang, Henan x1, Ma from Yuxi, Yunnan x2, Li from Dalian, Liaoning x1, Cai from Beijing x1, Jin 晋 from Tianjin x1, Zhang from Harbin, Heilongjiang x2, Wang from Zhoukou, Henan x1, Liu from Weinan, Shaanxi x2, Wang from Yantai, Shandong x1, F** from Guangzhou, Guangdong x1, Gong 贡 from Shijiazhuang, Hebei x1, Ye 冶 from Jiuquan, Gansu x1, Yang from Chengdu, Sichuan x1, Zhai from Tai'an, Shandong x1, Zhang from Shijiazhuang, Hebei x1, He from Cangzhou, Hebei x1, Zhao from Beijing x1, Nurmemet 努尔买买提 from Aksu Prefecture, Xinjiang x1)
L-M20 (TMRCA 14070 years) > L1-M22 (TMRCA 14070 years) > L1a-M2481 (TMRCA 14070 years) > L1a1-M27 (TMRCA 8860 years) > L-Y31961 > L-Y31214 > L-Y169295 > L-Z20087 (TMRCA 1310 years) 52 members (Jia from undisclosed x1, Zhao from Hulunbuir, Inner Mongolia x1, Luo from Linyi, Shandong x1, Hao from Baoding, Hebei x1, Deng from Xinxiang, Henan x1, Jin from Shenyang, Liaoning x1, Li from Shijiazhuang, Hebei x1, Li from Baoji, Shaanxi x1, Liu from Luoyang, Henan x1, Liu from Weinan, Shaanxi x1, Tu 图 from Kashgar Prefecture, Xinjiang x1, Ma from Luoyang, Henan x2, Liang 亮 from undisclosed x1, Lei from Xi'an, Shaanxi x1, Lu from Xuzhou, Jiangsu x1, Ding from Suqian, Jiangsu x1, Li from Shaoxing, Zhejiang x1, Yu from Xiangyang, Hubei x1, Li from Heihe, Heilongjiang x1, Li from Qiqihar, Heilongjiang x1, Run 润 from Jiuquan, Gansu x1, Zhang from Tianjin x1, Kang from Yulin, Shaanxi x1, Xiao from Bazhong, Sichuan x1, Shi 史 from Zhoukou, Henan x1, Ma from Haidong, Qinghai x1, Han from Zhoukou, Henan x1, Ma from Shuozhou, Shanxi x1, Zhai from Tai'an, Shandong x1, Lü from Xinxiang, Henan x1, Sun from Bengbu, Anhui x1, Nu 奴 from Kashgar Prefecture, Xinjiang x1, Ou from Suzhou, Anhui x1, Lan 拦 from Linxia Hui AP, Gansu x1, Mu 穆 from Fuyang, Anhui x1, Zhang from undisclosed x1, Luo from Tai'an, Shandong x2, Ma from undisclosed x3, Guo from Beijing x1, Deng from undisclosed x1, Liu from Jiaozuo, Henan x1, Chen from Yongzhou, Hunan x1, Wang from Ordos City, Inner Mongolia x1, Zhang from Binzhou, Shandong x2)
It appears that all known members of L-Y169295 may be from China. They seem to share a common ancestor within the last two (or perhaps three judging from the upper bound of YFull's 95% CI) millennia, but the phylogenetic position of the L-Y169295 clade is quite interesting: cf. https://www.yfull.com/tree/L-M27/. L-Y169295 belongs to L1a1-M27, which also includes the predominant branch of Y-DNA haplogroup L among Indians and Sri Lankans; however, Chinese L-Y169295 is a primary subclade of Southwest Asian L-Y31214 (TMRCA 8000 [95% CI 9300 <-> 6800] ybp according to YFull).
G-CTS1475 ≈ G-FT318152 (formed 8300 [95% CI 9700 <-> 7000] ybp, TMRCA 950 [95% CI 1650 <-> 375] ybp has three members on the current version of the YFull tree: YF065082 from Liaoning and YF065071 from Jilin under G-BY182126 (TMRCA 500 [95% CI 1000 <-> 225] ybp) and NA18549 (a Beijing Han) under G-CTS1475*.
ReplyDeleteOn 23mofang, G-CTS1475 has a total of 40 members with an estimated TMRCA of 8840 years, but 35 of those 40 members belong to the G-FT318152 subclade (TMRCA 2200 years), and 34 of those 35 share an additional SNP, MF204523.
G-PF3345 (TMRCA 9450 years) > G-CTS342 > G-Z724 > G-CTS1475 (TMRCA 8840 years) 4 members (Ying 滢 from undisclosed x1, Wang 王 from Qingdao, Shandong x1, Liu 刘 from Dalian, Liaoning x1, Liu 刘 from Changchun, Jilin x1)
G-PF3345 > G-CTS342 > G-Z724 > G-CTS1475 > G-Y177598 (TMRCA 2670 years) > G-MF280816 1 member (Liu 刘 Han Chinese from Linyi, Shandong x1)
G-PF3345 > G-CTS342 > G-Z724 > G-CTS1475 > G-Y177598 > G-FT318152 (TMRCA 2200 years) > G-MF295875 1 member (Cuan 爨 Han Chinese from Zhengzhou, Henan x1)
G-PF3345 > G-CTS342 > G-Z724 > G-CTS1475 > G-Y177598 > G-FT318152 > G-MF204523 34 members (Cao from Shijiazhuang, Hebei x4 [Han Chinese x1], Liu from Hinggan League, Inner Mongolia x1, Zhang from Xianyang, Shaanxi x1, Ma from Lanzhou, Gansu x1, Gao from Hengshui, Hebei x1, Liu from Qingdao, Shandong x1, Chen from Dingxi, Gansu x1, Dong from Xianyang, Shaanxi x1, Wang from Beijing x1, Wang from Linyi, Shandong x1, Yang from Linyi, Shandong x1, Niu from Lanzhou, Gansu x1, Zhang from Qingdao, Shandong x1, Yi 乙 from Suqian, Jiangsu x3, Liu from Handan, Hebei x1, Zhu from undisclosed x1, Li from Yantai, Shandong x2, Zhang from Beijing x1, Liu from Linyi, Shandong x1, Li from Binzhou, Shandong x1, Zhang from Yantai, Shandong x1, Zhang from Jining, Shandong x1, Liu from Tai'an, Shandong x1, Liu from Yancheng, Jiangsu x1, g** from Xi'an, Shaanxi x1, Bai from Xi'an, Shaanxi x1, Liu from Chengdu, Sichuan x1, Liu from undisclosed x1)
Peter Frost's ideas are like almost all evo-psych, just bullshit. If his paleolithic European ancestors were selecting for greater physical beauty then one has to wonder how he ended up looking like he does..
ReplyDelete@Vasishta
ReplyDelete"It is irrelevant, the much later Hittite period samples show no extra ancestry than what I have already described. Isparta EBA, central Anatolia (2400bce) also lacks your ancestry."
There is very little doubt that Hittites were intrusive in central Anatolia and started to rule over a Hattian population. Members of the royal family the northern branch bore Hattian names and parts of the prayers were done in the Hattian language. Furthermore, Hittites cremated more often than not and no royal inhumation is known, so there is a large chance we will never find any useful Hittite royal DNA.
Hence you can't conclude anything from the Damsgaard samples, just like absence of any Greek admixture in Ptolemaean age mummies in Egypt means anything at all with regard of the origin of the Ptolemaeans.
Maybe we should be charitable and interpret Andrzej as talking about his a(utosomal) dna?
ReplyDelete@gL
ReplyDeleteHow do you explain this PCA, which obviously shows Germanic influence in Estonia that is missing in Latvia and Lithuania?
https://blogger.googleusercontent.com/img/a/AVvXsEgk8wTciXnA-HPQvdsq9H9z-Ex-nbObbdfk-FN3Tvrm-u2hLu_vJlt-hKo5863XEYuLHVCZTJQGvXKp0pJ8bbhe7t0A0AsKRRvPbffuME3Uw10K6ajQ0pwYV69Y2LpteZJ-RRJU3VjjfaTI63VgsvssLLdppW7MnPZONcp7X-FXTaKExd7Q2fSzGUI4JQ=s1000
@ epoch
ReplyDelete“ Hence you can't conclude anything from the Damsgaard samples”
In the damagaard paper suppl it says they’re Assyrian colonists killed in a market place, or something like that
So they don’t actually have anything to do with Hittites
Curiously, Hatti land was invaded from the south, although the IEs ultimately came from the west
@gL
ReplyDelete"There is absolutelly no relation to Germans in "Estonian Germans", because their origin(and dna) is Estonian - they switched to German language, because that was language of ruling class."
Did you see a lot of Estonian German DNA or what evidence do you have? Your explanation at least sounds dubious. For if they switched to German because it was supposedly the language of the ruling class, then why are there not more of them? Why didn't Estonian go extinct? As far as I know, the thing that's different about Baltic Germans (i.e. Latvian and Estonian Germans), as compared e.g. to East Prussian, Pomeranian or Silesian Germans, is that in the Baltic there was never any rural colonisation through German peasants. The Baltic Germans were exclusively townsfolk of the middle and upper class. And as such they were more mobile and had wider contact networks. But I suppose it also means that they were always a small minority.
BTW, Estonia belonged to the Teutonic Order state from the 13th century until 1561, then it became Swedish, and in 1721 Russian.
ReplyDeletegL is full of shit.
ReplyDeleteThere were Swedish speaking settlements in western Estonia until very recent times. Then there were also the Vikings, Germanic Crusaders, etc.
Completely anecdotal but green eyes seem especially common in southern Finland. A class of 30 I attended did a count with 16 green, 9 blue and 5 brown with three of the browns foreigners. Blue eyes are definitely a lot more common in Estonians, its something that does stick out even to Finns.
ReplyDeleteI wouldnt be surprised if the association with green eyes and Siberian is simply explained with Finnish admixture carrying both.
@Gaska
ReplyDeleteKurganist? What the hell is a kurganist. Sounds like a Norwegian black metal band. VK531 (2559-2290 calBCE) is R1b-Y13200>Y13202* (check YFull). He has nothing to do with WSHs, M269, or L51. Radboud over at Anthrogenica, had this to say about VK531’s autosomal profile: “VK531 is similar to the Combed Ware (Pit-Comb Ware culture) samples from the Baltic region. These specific samples also cluster between SHG's and EHG's.” Mittnik et al. (2018): “Using the qpWave/qpAdm framework, we modelled the Baltic Mesolithic hunter-gatherers as a two-way mixture between EHG and WHG (Fig. 3), which reveals a difference in mixture proportions between the more northern individuals from the Latvian site (65–76% WHG with 24–35% EHG; Supplementary Table 3) and the samples from the Lithuanian sites to the south (88–100% WHG with 0–12% EHG; Supplementary Table 3)…Using qpAdm, we confirm the previously published result of SHG being formed by admixture of WHG and EHG (57 ± 2% WHG with 43 ± 2% EHG)…Both EHG and SHG share a non-negligible component in ADMIXTURE analysis that is maximized in some modern Native American populations which points towards ANE ancestry, as represented by the MA1 and AG3 samples from Palaeolithic Siberia (maroon component in Fig. 2a, Supplementary Fig. 4). Indeed, D-statistics show that EHG and SHG share significantly more alleles with MA1 and AG3 than WHG…Similarly to the Baltic Mesolithic, the later Eastern Baltic Neolithic hunter-gatherers of the Narva culture exhibit varying proportions of EHG (0–46%) and WHG (54–100%) ancestry (Fig. 3 and Supplementary Table 3)…The later individuals attributed to the Baltic MN CCC exhibit a significantly higher affinity to EHG with the ancestry proportion estimated at 68–99% EHG and 1–32% WHG (Fig. 3, Supplementary Table 3).” Lamnidis et al. (2018) found the following: 63.3% EHG and 19.8% Yamnaya for CCC/Pit-Comb Ware, 45.5% EHG in two PWC Sweden HGs, 40.8% EHG in two SHGs, 15.5% EHG in two Narva HGs, 5% and 17.2% EHG in two Kunda HGs, 19.7% and 26.2% EHG in two Latvia HGs, 20.7% EHG in a Latvia MN sample, and 31.5% Yamnaya in another Latvia MN sample. Jones et al. (2017), and Matthieson et al. (2018) got practically the same results (30% EHG in Kunda, Narva, and 65%-100% EHG in CCC). VK531 was like the SHGs and the CCC HGs; he had a significant amount of EHG ancestry, and that’s where his R1b-M73 comes from. Additionally, VK531 lived way after Srendy Stog, Khvalynsk, Vonyuchka, Progress, Repin, Yamnaya, Afanasievo, and Corded Ware.
The earliest R1b-M269 sample we have—Smyadovo I2181—also has EHG/steppe ancestry, from MathiesonNature2018: “In two directly dated individuals from southeastern Europe, one (ANI163) from the Varna I cemetery dated to 4711-4550 BCE and one (I2181) from nearby Smyadovo dated to 4550-4450 BCE, we find far earlier evidence of steppe-related ancestry (Figure 1B,D).” When they modeled him in D stats (Mbuti.DG, CHG, Balkans_Chalcolithic, Balkans_Chalcolithic_outlier) they found the following: “Varna_outlier has Steppe ancestry. Balkans_Chalcolithic_outlier may have steppe ancestry but has no evidence of CHG component (however number of SNPs is low).” In the paper’s supplement, I2181 has 46.1% Yamnaya in qpAdm, with a standard deviation of 17.4%. In admixture analysis, he picked up EHG, and a larger Yamnaya_Samara component. I2181 and the Varna outlier are both on the Khvalynsk cline, unlike the other Balkans Chalcolithic samples: https://3.bp.blogspot.com/-gpo-jpYFsY0/WST-FhAWHMI/AAAAAAAAFpU/VISsvervHfQ34fSBLM7c1KOIkLMAxxhTwCLcB/s1600/Steppe_clines.png
@Gaska
ReplyDeleteHere’s data from Patterson et al. (2022), Supplementary Tables, S5: I2611 has 59.9% Steppe autosomal DNA, with a 2.5% margin of error; I3019 has 44.2% Steppe with a 4.2% margin of error; I3035 has 46.5% Steppe with a 2% margin of error. You’re also being disingenuous. I’ve told you before, CLL007 has not been directly carbon dated (and calibrated). The highest C14 range for an R1b-M269 sample in that paper is 2296-2060 BCE (sample ID: MDP001). Two Iberian CA samples—CDM001 and CDM002—have yielded C14 date ranges of 2550-2234 BCE and 2579-2346 BCE respectively. This is despite them being archaeologically/indirectly dated to 3300-2300 BCE. As I told you before, just because a sample is archaeologically dated to a certain period, doesn’t mean its from that period, nor does it mean it occupies the median or high end of the archaeological age range. CLL007—like the aforementioned two samples—could give a C14 date that corresponds to the low end of the age range (2500-2200 calBCE), which would correlate with the initial dispersal of steppe ancestry into SW Europe. CLL007 also has Steppe autosomal DNA: “CLL007 (Arc. ID: Lech 11 (LE11B)): Adult male burial. This individual was considered intrusive and thus excluded from the analysis, as it showed steppe-related ancestry.”
The Swiss samples. MX304 has 21.5% Yamnaya_Samara (SD is 14.6%), and MX310 has 30% Yamnaya_Samara (SD is 16.9%) according to the authors of the study. Additionally, MX304 is positive for R1b-P311. Check Figure 2 and Supplementary Data 4. They have Steppe ancestry, but they also have a very low number of SNPs (29434 and 28525 respectively). XN191 belongs to R1b-L754, and he has 20283 SNPs, which is abysmally low coverage. So no link to M269 there either. Furthermore, contemporaneous pre-Steppe R1b males (from Germany and Czechia), all belong to V88 (sample IDs: I14169, I14176, I14173, I0559, I1593).
@Gaska
ReplyDeleteYou and goddamn ATP3, is it 2015 again? ATP3 (3516-3365 calBCE) is a very poor quality sample. He has an abysmally low SNP count of 37166 (0.031 autosomal coverage), along with some damage and/or contamination (Z=2.3 contamination based on damage/non-damage comparison). Regardless, hypothetically speaking, the Y-DNA SNP calls can still be correct. I6222 has consistent ancestral and derived calls, despite being damaged/contaminated. He belongs to R1b-L52*/P310*; there were no further calls for anachronistic, descending clades. Such a case would indicate contamination. Additionally, it’s practically impossible for a modern male to have P310*. Genetiker found the following SNPs in ATP3, and all of them demonstrate damage and/or contamination: A00-AF12, E-M78-L547/PF2166/V4057, J2-Z6046-SK2198/S18476, F-Y1811/FGC2054, GHIJK-M3773/CTS12673, F-PF2756, K-CTS9278/PF5501/M2693, O-F1835, Q-Y1109, Q-YP1618, Q-Y18433, I2-Y16441-Y16447 (formed 2100-1800 BCE), R-Y479/F370/PF6047/M708, R1-M748/YSC0000207, R2-Y3545/FGC24163, R1b-P297-Y97/FGC46, and R1b-L21-Y16874-Y17204 (formed 1500 BCE). Let’s break this down; parentheses indicate clades that are SNP consistent.
PF2756 is a SNP in numerous haplogroups, including—but not limited to—J2-M102, J2-PF5008, J2-PF5160, J2-BY186473, J2-Y64860, (J2-Z6046), J2-Z6064, J2-CTS4800, J2-Y7687, F-Y27277, J1, (I, K, G, H), and E-M250. According to YFull, M3773/CTS12673 is a SNP in (F), C-Y138372*, and A-FT174073, not GHIJK, while CTS9278/PF5501/M2693 can correspond to (J-FGC61612*, G-PF3177*, K, I-CTS10057, I-BY33169*), I-BY138*, (I-PH5234), and I-BY3605*.
Furthermore, ATP3 had PF6401, which is a SNP found in the following clades: A00*, E-Y175024, E-Y45797, E-L241*, C-V20*, C-FT59486, C-Y10492, C-M208*, C-Y507, C-Y11606*, G-Z36520*, G-FGC34434*, G-PF3177*, G-BY133179, G-Y144865, G-Z6128*, G-BY146155*, G-Z1903*, H-P96*, H-Y19962*, I-FGC21819, I-F1295*, I-FT344596*, I-FT344665*, I-FT344602, I-Y28222*, I-FTA37476, I-BY33164*, (I-BY33169*), I-Y79427*, I-L1498, I-FT27731*, I-A8742*, I-BY37286*, I-BY60222*, I-Y4882*, I-Y16473, I-BY193138*, I-S2555*, I-Y52134, I-Y34539*, I-Y7240, I-BY138*, I-Y43545, I-Y3709*, I-FT380000, I-BY166007*, I-BY168618*, I-Y3712*, (I-Z161*, I-PH5234, I-L801*), I-BY3605*, J-Y159482*, J-FTA1458*, J-MF10501, (J-FGC61612*), J-Z36829*, L-Y6288, T-Y13280*, T-Y63197*, (T-Y261958), S-B255*, Q-Y144860*, (Q-FT380500), Q-Y2700*, Q-F2019*, Q-BZ4911*, Q-BZ2222*, Q-Y12449, Q-FT421589*, Q-YP4547, Q-Y4276, Q-Y4277*, Q-M848, R-M459*, R-BY149647*, R-S23592, R-YP1558, R-YP1558*, R-Y874*, R-Z283, R-Y126934, R-Y2395, R-YP986*, R-Z280, (R1b), (R-P297), (R-FTA35718, R-FTA35720, R-Y127541, R-PF6287*, R-PF6362), R-Y3370*, O-K2*, (O-MF15397*)
The other SNP is PF6518, which is found in the following: A00*, C-V20*, C-FT59486, H-Y21612*, I-FT344596, I-FTA37476, (I-BY33169*), I-Y169013, I-FT33727*, I-BY138*, I-Y3712*, (I-CTS10057, I-PH5234), J-MF10501, (J-FGC61612*), L-Y31961*, (T-Y261958, Q-FT380500), Q-Y4943a*, R-Z2123, R-Y88962*, (R1b), (R-M269), (R-FTA35720), (O-MF15397*)
@Simon Stevin
ReplyDeleteKurganist is someone who only has steppe (or percentage of steppe ancestry) in his brain and is incapable of recognizing his mistakes.Using the percentage of steppe ancestry to prove the origin of R1b-L51>L151 (or IE language) in the steppes is the last resort of desperate people who have no other arguments. Everything would be much easier if you would present convincing evidence, i.e. some case of this lineage in Yamnaya, Sredni Stog or in any other steppe culture.
1-You can go on thinking that I6222 is the solution to your problems, I have already told you that you should ask for independent opinions because that sample is simply contaminated garbage. Nevertheless it is amusing to see how you defend the existence of P310 in Mongolia because if that were true, that means that it would be linked to the Afanasievo culture that is to say to the Yamnaya culture. You should explain it to MaxPlanck who just put in writing that it is impossible that the Yamnaya culture is the origin of that lineage because of the obvious discontinuity in the male uniparental markers with the CWC and BBC. In this case, I think they are right and that the Yamnaya ship sailed a long time ago.
2-ATP3 has been the main topic of discussion for 7 years. Kurganists have denied that it was M269 from the beginning and many people have been banned in different forums for advocating otherwise. This is how the Kurganists have operated, exactly like the Holy Inquisition. Finally when it appeared published in the Reich data it was recognized at least as P297. BUT R1b1a/2-M269:ATP3-PF6518-Marker PF6518 (Y: 23,099,729 A>C) is a transversion, and it is not a C>T or G>A which could be the result of DNA damage. Therefore, this could be a case of some kind of R1b1a/2-M269 being present in the Pre-Bell Beaker/Chalcolithic (5,526-5,372 ybp-Cal BP + 60 years), ergo we were right.
3-CLL007-I have already explained that it was found in a typical Mediterranean Neolithic site, I have sent you the grave goods that were found and the burial ritual. It has absolutely nothing to do with the BB culture and the controversy will end when they date the sample again (someone has told me that it will be around 3.000 BC), it will be fun to see your face when they confirm it officially.
4-Tell your friends at anthrogenica that in Iberia we also have the oldest known DF27.-EHU002 El Hundido, Burgos-(2.434 BC) belongs to R1b1a/1b1a/1a2a/7 positive A12032/S24844
They have been talking nonsense about Quedlinburg for 7 years to have everyone fooled.