search this blog
Sunday, May 3, 2020
Understanding the Eneolithic steppe
Archeologist David Anthony has teamed up with Harvard's David Reich Lab to work on a paper about the Eneolithic period on the Pontic-Caspian steppe.
A couple of other labs are also preparing papers on similar topics, and they've already sequenced and analyzed many of their ancient samples (for instance, see here). However, I don't have a clue when these papers will be published. My guess is that we'll have to wait a year or so.
Needless to say, knowing what happened on the Pontic-Caspian (PC) steppe and surrounds during the Eneolithic is crucial to understanding the origins of the present-day European gene pool. It's also likely to be highly relevant to the debate about the location of the Proto-Indo-European (PIE) homeland.
In this blog post I'll explain what I've learned about the Eneolithic peoples of the PC steppe based on already published data.
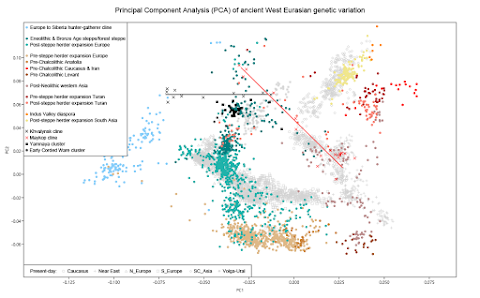
If we ignore Steppe Maykop samples, the currently available Eneolithic individuals from the eastern part of the PC steppe form an essentially perfect cline in my Principal Component Analysis (PCA) of ancient West Eurasian genetic variation.
The cline runs from the Mesolithic hunter-fishers of the Eastern European forest zone to those of the Eneolithic sites of Progress 2 and Vonyuchka in the North Caucasus foothills. Let's call this the Khvalynsk cline, because three of the samples are from a burial site in the Volga River valley associated with the Khvalynsk culture. The relevant datasheet is available here.
The reason that these samples form the cline is because they carry different ratios of admixture related to Caucasus hunter-gatherers (CHG) from what is now Georgia. Moreover, the Khvalynsk individuals appear to be relatively recent mixtures between sources rich and poor in this type of ancestry.
I also marked a Maykop cline on the plot. This cline is made up of individuals associated with the Maykop and Steppe Maykop cultures from the Caucasus Mountains and nearby parts of the PC steppe, respectively. The Maykop culture is dated to the Early Bronze Age (EBA) period, but the PC steppe was still part of the Eneolithic world at the time.
The Maykop cline is more complicated than the Khvalynsk cline, because some of the Maykop individuals carry genetic components that the others lack. These genetic components are closely related to the aforementioned CHG, as well as Anatolian Neolithic farmers (ANF) and Western Siberian hunter-gatherers (WSHG).
Note that the two clines intersect, but this isn't because any of the Khvalynsk cline samples harbor Maykop-related ancestry. It's largely because the Steppe Maykop individuals carry high levels of Vonyuchka-related ancestry.
So unless we're dealing here with a remarkable string of coincidences, then the Vonyuchka hunter-fisher must be a decent proxy for the people who spread significant levels of CHG-related ancestry north of the Caucasus.
The important question, therefore, is where and when exactly did this population form? And it's a question that the authors of the aforementioned upcoming papers should be aiming to answer comprehensively.
In my view, it was the result of interactions between the hunter-fishers of the North Caucasus and the southernmost parts of the PC steppe during the Neolithic period, perhaps around 6,000 BCE, just before significant ANF-related ancestry spread across the Caucasus during the Eneolithic. That's because the Progress 2/Vonyuchka samples lack ANF-related ancestry, or at least an obvious signal of it, and are dated to ~4,200 BCE. And when I say Neolithic in this context, I don't mean the Near Eastern type of Neolithic with well developed farming, but rather the local type of Neolithic still based on hunting and fishing.
Now, obviously, the people of the Corded Ware and Yamnaya cultures were the children of the Eneolithic PC steppe. So you might be wondering how they fit into all of this. I still don't know, and apparently neither do the scientists at Harvard (see here). However, I'd say that the Maykop cline isn't relevant to this question. The Khvalynsk cline might be relevant, but even if it is, this doesn't necessarily mean that the Yamnaya people are by and large derived from the Khvalynsk people.
Here's the same PCA plot as above, but this time with early Corded Ware and Yamnaya samples also highlighted. Note that, apart from a few outliers, they form a rather tight cluster that is shifted slightly away from the Khvalynsk cline, but probably not in the direction of the Maykop cline.
A couple of the Yamnaya outliers are shifted towards the "eastern" end of the Khvalynsk cline, and thus near the Progress 2/Vonyuchka samples. This isn't surprising because these Yamnaya individuals are from burial sites close to the North Caucasus and probably harbor significant levels of local ancestry.
The most extreme Yamnaya outlier, from a site in what is now Ukraine, is clearly shifted towards the Maykop cline, and even towards the Caucasus Maykop cluster. However, this is a female with no grave goods and she may have been a foreign bride or captive, possibly from a late Maykop settlement. It's also possible that her 3095-2915 calBCE dating is wrong.
I'm pretty sure that when we find out why the Yamnaya cluster is so deliberately shifted away from the Khvalynsk cline, we'll also discover how the early Corded Ware and Yamnaya populations formed. For now, I strongly suspect that this has something to do with gene flow from the western edge of the PC steppe and the ethnogenesis of the Sredny Stog culture, which was located just west of the Khvalynsk culture.
By and large, the PC steppe is still seen by historical linguists and archeologists as the most sensible place to put the PIE homeland.
However, a theory that the PIE homeland was located somewhere south of the Caucasus, and that instead the PC steppe was the late or nuclear PIE dispersal point, has gained popularity in recent years, largely thanks to the apparent lack of PC steppe ancestry in a handful of samples from Hittite era Anatolia. In this scheme, the Maykop culture took PIE into Eastern Europe and the Yamnaya culture subsequently spread late/nuclear PIE from the PC steppe, while Proto-Anatolian, the ancestor of Hittite, was introduced into Anatolia from the east along with Maykop-related ancestry.
This is possible, in the sense that almost anything is possible, but it doesn't strike me as the most parsimonious interpretation of the facts.
Even before ancient DNA, it was known that the Maykop culture colonized parts of the PC steppe, at least for a short time, and probably had contacts with the Yamnaya people and/or their antecedents. But it was generally seen as the vector for Caucasian and other non-Indo-European influences in PIE.
Moreover, not only were the Maykop and Yamnaya populations of fundamentally different genetic origins, but apparently the Yamnaya people didn't absorb any perceptible Maykop ancestry as they expanded into the North Caucasus region at the tail end of the Maykop period.
That's really difficult to explain if we assume that these groups were close linguistic relatives, and much easier to reconcile with the assumption that they were derived from different worlds culturally and linguistically.
Another important question is what happened to the Steppe Maykop people, because right now it looks like they vanished almost without a trace, essentially as if they were pushed out or even erased by the Yamnaya expansion. If they were indeed pushed out or erased, then it's likely that their language was as well.
As for the lack of PC steppe ancestry in Hittite era Anatolians, I honestly can't see this is as a significant obstacle to a PIE homeland on the steppe, especially if we consider that the most widely accepted Indo-European phylogenies show the Anatolian family as the most basal node.
In the opinion of the vast majority of experts, it's the most basal node because the Proto-Anatolian speakers were the first to leave the PIE homeland. And if they were indeed the first to leave the homeland, then why should we expect their descendants to harbor significant ancestry from the homeland? In my view, such an assumption would contradict the most widely accepted Indo-European phylogenies.
See also...
The Caucasus is a semipermeable barrier to gene flow
Subscribe to:
Post Comments (Atom)

445 comments:
«Oldest ‹Older 401 – 445 of 445@epoch
In fact, this simple 2 way model does as well your wholly implausible model with ancient sources.
Target: RUS_Khvalynsk_En_scaled:I0434
Distance: 3.0883% / 0.03088285
77.4 RUS_Khvalynsk_En
22.6 TJK_Sarazm_En
@Davidski
I was trying to "triangulate" CHG, Hotu HG, and the various Iran Neolithic samples with various references form the surrounding regions with G25 nMonte. The fits are all horrible, the right ancestral populations are obviously missing, but I think it still gives useful information for comparison. The results probably won't surprise you.
1. Just a footnote, but both ancient Iran and CHG absolutely "refuses" anything WHG related, until the extent that Pinarbasi is down to zero in the presence of Barcin_N. But this is a common trait of CHG and Iran, so not so interesting here.
2. All the Iranian samples have a lot of ancestry from possibly multiple southern population that is missing from CHG. And they are also missing from Yamnaya. The differential affinity to Natufians and Onge (Onge in the absence of anything ancient Indian that is not also Iran admixed) is so big that there is no way it would not turn up in Yamnaya if Yamnaya had the bulk of its "Caucasus-Iran" ancestry from actual Iran Neolithic.
I know (and I apologize again) that I am not using the right tool, but still, let's say we take these results on face value. Based on this there is no way Yamnaya had ancestry from anywhere south of the South Caspian coastline in less than 6000 years time depth (that is 6000 years before Yamnaya) the very least.
Just for general interest, there are the G25 nMonte results:
"sample": "GEO_CHG:Average",
"fit": 24.4429,
"Anatolia_Barcin_N": 51.67,
"RUS_MA1": 45.83,
"Onge": 2.5,
"Levant_Natufian": 0,
"sample": "IRN_HotuIIIb_Meso:Average",
"fit": 20.9982,
"RUS_MA1": 54.17,
"Anatolia_Barcin_N": 19.17,
"Levant_Natufian": 19.17,
"Onge": 7.5,
"sample": "IRN_Ganj_Dareh_N:Average",
"fit": 25.1705,
"RUS_MA1": 43.33,
"Levant_Natufian": 26.67,
"Anatolia_Barcin_N": 18.33,
"Onge": 11.67,
The other Iran Neolithic groups are essentially the same, so I won't list their results.
"sample": "IRN_HotuIIIb_Meso:Average",
"fit": 13.6151,
"GEO_CHG": 61.67,
"RUS_MA1": 21.67,
"Levant_Natufian": 8.33,
"Onge": 8.33,
"sample": "IRN_Ganj_Dareh_N:Average",
"fit": 16.2991,
"GEO_CHG": 73.33,
"Onge": 13.33,
"Levant_Natufian": 10,
"RUS_MA1": 3.33,
"sample": "Yamnaya_RUS_Kalmykia:Average",
"fit": 8.4324,
"RUS_AfontovaGora3": 42.5,
"GEO_CHG": 33.33,
"Anatolia_Barcin_N": 14.17,
"ITA_Grotta_Continenza_Meso": 10,
"Levant_Natufian": 0,
"Onge": 0,
"RUS_MA1": 0,
Yamnaya comes out with total zero ancestry from all the references that differentiated Iran from CHG in the comparative "triangulating".
@Slumbery
I got further interested in this topic, do you have any good model for EHG?
@Davidski
Away from desktop and R, but Yamnaya seem to take 20% Steppe_Maykop in Vahaduo, with the rest being EHG, CHG and Globular_Amphora with d=0.0537. It doesn't really seem to need it though, if it is excluded, distance only increases to 0.058.
@Matt,
Fair enough. Recent drift is important to modern variation. But, in G25 PCA, East Europe is only location in West Eurasia with strong recent drift.
As rebuttal, I will say West Eurasia PCA more than anything is determined by Paleolithic ancestry ratios. In terms of overall European and West Eurasian variation, Paleolithic drift plays the biggest role.
But, yes I was too simplistic in what I said about Paleolithic drift being much more important.
@Slumber
I suggest you go through published genomic papers about it, there are several out there that is pointing to what i'm saying. I'll link this maykop one here for example. https://www.nature.com/articles/s41467-018-08220-8
EDAR allele presence in EHG, SHG and Maykop all points to what i'm saying in my previous posts. This allele is not found in ANE. It's coming from actual East Asian ancestry.
"This is exemplified by the more commonly East Asian features such as the derived EDAR allele (Supplementary Note 7), which has also been observed in HG from Karelia and Scandinavia. The additional affinity to East Asians suggests that this ancestry is not derived directly from ANE."
"Pure" ANE did not arrive to Eastern Europe, it arrived as combination of high ANE + low East Asian (via Baikal) which came with steppe cord ceramics and microblade technology, found in all EHG admixed sites. Lazaridis suggestion in Dzudzuana study makes sense archaeologically as well with 13kya arrival.
East Asian/EDAR allele present in EHG was likely diluted by eneolithic period by mixing with other components. In contrast, ABCC11 allele was likely present in both ANE and East Asian at high frequency, allele carried well into bronze age steppe, that's why it still has good presence in Eastern and Northern Europe.
Thank you, Garvan. Much appreciated.
@Samuel Andrews
Thank you for the detailed reply, Samuel. I was getting a handle on the concepts of founders effect and bottlenecks, but wasn't sure what shared drift referred to. If I understand your description correctly, drift is describing how a population has differentiated itself from another one. By determining which populations share drift, we can determine whether or not they branched off from a common ancestor, and we would be able to proximally estimate how long ago this occurred? When you say Finns are more drifted than Hungarians, Finns due to time and isolated reproduction have differentiated themselves away MORE than the Hungarians, relative to other Europeans?
@RobertN,
It means Finns are more related to other Finns than Hungarians are to other Hungarians. An important fact about population genetics is some populations are more drifted than others. All populations represent genetic groups but some are more genetically-defined than others.
@Sam, following partly a response to you (not rebutting anything in your comment so don't take it that way!), and partly some general Global 25 modelling geekery that could be of interest to yourself anyone who does a lot of that using Vahaduo (@Arza, etc.), or is interested in the overall fit of ancient samples to modern pops on G25. Mainly stuff I found out while messing around with modelling yesterday, prompted by your discussion:
Sure, yeah, for sure, West Eurasia PCA is dominated by these components that split in these ancestry at these sort of dates (late Upper Paleolithic / early Mesolithic ancestry).
In hindsight this does make perfect sense (I thought back in like 2013-2014 more of the West Eurasia PCA position would be dominated by recent drifts, but in hindsight it makes sense that it would be dominated by these).
In terms of drift in Europe that is not modelled by the 3-4 components (Barcin, Steppe, WHG, Nganasan) approach, it actually seems really hard to work out what is recent drift and what is just unmodelled long term structure where actually the real admixing populations in different regions were slightly more different than we'd think using a few references.
But if it is drift, for sure the local drift effect is probably something like 3x or more as strong in NE Europe as NW Europe. Though there still is a region drift effect in NW Europe, that comes when we look at comparing models to reality.
Here's an illustration, some Vahaduo fits where I'm using a lot of groups to really go all out and try to capture all the drift dimensions in a set of West Eurasian pops (inc. Baltic_BA to capture the NE European variation): https://imgur.com/a/WGAJXt3
First set of results is in alphabetical order, second is in fit order of increasing fit quality (worse->better). The most least fitting populations are Volga-Ural ones, which is because I haven't included a Volga-Ural reference (no FIN_Levanluhta, etc). Then the NW Europeans also come in after that.
So there is some unmodelled difference there - I didn't find that including likely NW representative ancient dna like Roman_Iron_Age or FIN_Levanluhta_o etc actually helps too much here, strangely enough, and it just seemed totally unexplainable with any adna I could find...
Another way to see this is to put the models on a tree with the real populations: https://imgur.com/a/EcEGhRz .
Most models from my Vahaduo fits "twin" as a parallel branch with their true population, suggesting its a fairly good model within limits, but there's some parallel branching in NW Europe and Russia Volga-Ural in particular (which suggests the model is a bit imperfect).
Another third way to see it is to use the approach of reprocessing PCA that we invented on here, and which forms the basis of Vahaduo Custom PCA, with the model components and their target populations together: https://imgur.com/a/WqKTl5p .
We can see that when PCA together real populations (red dots) and models (blue dot) are pretty close to each other, particularly in main dimensions dominated by ancient components, but some diverge more than others, eventually (and these are the same NW European and Volga-Ural that pop out on the tree above).
So there is still some unmodelled drift.
(cont from previous post):
One final more general point about the actual comparison of models to reality, that's not much relevant to the questions about European drift, but which is relevant generally to modelling modern people with ancient dna on Global 25.
As well as doing reprocessed / custom PCA with the models using ancients, and the real populations, we can just look at them in the normal G25 dimensions together: https://imgur.com/a/4z1lQxR
You can see that there is a structure where the model fits (blue dots) are parallel and very slightly compressed towards the centre of the plot. If we look at the position of the source populations, it's immediately obvious why this. The blue model dots are "jogged" slightly towards the centre of the plot.
It looks like although the G25 is probably the best model yet for dealing with shrinkage and compression of ancient dna on PCA, there is still some very small shrinkage.
This means that we probably won't ever get totally perfect models with G25 adna for moderns that are quite as good with modelling moderns with other moderns, or ancients with ancients...
It may not matter as much for using all adna together as sources, because it looks like all adna are compressed roughly equally (so effect cancels). But comparing fits distance of moderns with ancients to fits of ancients with ancients or moderns with moderns may be not a fair comparison.
Anyway, I think this strategy of comparing a mass set of models to real populations has some pretty useful insights, in situations where the adna's good enough to allow it. It might allow us to quantify shrinkage rather than just having the impression that there is some, or there isn't much.
@Matt
You're right that there's some shrinkage in the G25 that affects the ancestry proportions. For instance, the hunter-gatherer ancestry proportions are somewhat inflated for Eastern and Northern Europeans.
The problem is especially pronounced when using distal sources, but it almost disappears with more relevant sources that probably share specific drift with the modern samples.
Also, high quality modern samples sequenced with the capture method seem to produce very crisp results, but I've only been able to test that with a handful of individuals, and they're not in the G25 datasheets yet.
Well, to be clear, I'm not actually arguing it necessarily affects the proportions (because I'm thinking all ancients equally shrunk and equally imperfect), just the overall minimum distances achievable.
Re; proximal sources, how proximal are we talking? Is it like, if used Beakers rather than CW_Early+Europe MN for NW Europeans, no compression in models on dimension 1+2 compared to reality? Or even more proximal than that? Baltic BA seems to show quite a bit of shrinkage in dimension 1 and 2, not less really than Barcin it didn't seem. I'll give that a try to see if 1+2 shrinkage is less pronounced with Beaker as a source.
@Vincent
" It's coming from actual East Asian ancestry."
What's your point here? That Yamnaya had East Asian admixture?
"EDAR allele presence in EHG, SHG and Maykop all points to what i'm saying in my previous posts. This allele is not found in ANE. It's coming from actual East Asian ancestry."
You don't know if it was present in ANE or not, we only have a few ANE samples. It was not present in all EHG populations either IIRC.
@Matt
I meant this sort of thing. But I think this is acceptable considering that we don't yet have the right steppe population that contributed the ~50% of Yamnaya-related ancestry to Poles, nor the most relevant Neolithic farmer population (which is probably why the fits are poor). qpAdm does a more stable job with distal proxies, because obviously it runs on formal statistics.
G25/Vahaduo
Target: Polish
Distance: 5.2282% / 0.05228229
42.0 Yamnaya_RUS_Samara
34.4 Anatolia_Barcin_N
21.4 Baltic_LTU_Narva
2.2 RUS_Karelia_HG
qpAdm
Polish
Yamnaya_RUS_Samara 0.479±0.042
Anatolia_Barcin_N 0.324±0.016
Baltic_LTU_Narva 0.174±0.013
RUS_Karelia_HG 0.022±0.034
Full qpAdm output
Yeah, I'm not saying the models are bad, or proportions are wrong, just that:
a) if there are systematic, very slight, shrinkages among adna, reasons why adna fits for moderns may not be quite as perfect (close to zero) as modelling moderns with moderns for ex, or ancients with ancients. There might be fundamental limits to how close the fit of moderns with adna can get, even with perfect references, just because of properties shared by adna that make them shrink a little on this method. (This is analogous to the sort of problems in qpAdm that were in Harney's paper).
b) looking at plots of big sets of fitted models vs real populations and seeing how they *systematically* differ in position might be a useful way to quantify how much shrinkage is present in a plot. Because if it's low and we're just eyeballing it, we might not see a low level.
@davidski
In a previous post comment you had claimed that steppe_en is ancestral to khvalynsk. do you still hold that claim after knowing that khvalynsk is definitely older than steppe_en?
As far as the Q1a2 in Khvalynsk is concerned, all papers consider it to be central asian in origin.
eg. this 2019 paper https://bmcbiol.biomedcentral.com/articles/10.1186/s12915-018-0622-4 has 6 Q-M25 and subclade samples out of 151 Q samples. 3 are from uzbekistan, 1 from lorestan iran, 1 from pakistan, 1 from India. None from siberia, americas, europe, east asia or alaska. The M25 in ancient east siberia and beringia did not survive.
"Indeed Q-M25, which is frequent in modern Western Eurasians [53, 54], is described as Q-L712 and Q-L713 in ancient samples from the Beringian area [33] indicating that, during the warmer mid-Holocene period, populations carrying different haplogroup Q lineages reached the former Beringian area but gave a limited contribution to the modern Y chromosome gene pool."
Q-M25 does not look like a turkic language marker. Only 1 in 189 modern native altaians was positive for Q-M25 https://www.sciencedirect.com/science/article/pii/S0002929711005490#app2
This is a total list of all modern Q-M25 and subclades so far from various papers, Yfull, etc. (compiled in Grugni et al 2019)
Mongolia 1
Uzbekistan 4
Lebanon 2
Caucasus 1
Altai Republic 1
Bulgaria 2
Iran 27
Iraq 1
Mongolia 3
United Kingdom 1
Hungary 1
Poland 1
Russia 3
Pakistan 1
Turkey 1
Bahrain 1
India 1
from another 2019 paper https://europepmc.org/article/pmc/pmc6851379
Hg Q1a2- M25 is very rare in Europe, where it has highest frequency among Seklers (a Hungarian speaking ethnic group in Transylvania) according to Family Tree DNA database. Ancient samples with Hg Q1a2- M25 are known from the Bronze Age Okunevo and Karasuk cultures, as well as Middle Age Tian Shan Huns and Hunnic-Sarmatians implying possible Hunnic origin of this lineage in Europe, which is confirmed by the Hg of our Hun/1 sample, derived from Transylvania. One Conqueror sample KEF1/10936 belongs to Q1a- F1096, possibly to the Q1a1-F746 sister clade of Q1a2- M25, which was not tested in our experiment. Q1a1 is present in East Asia; Mongolia, Japan, China, Korea and its presence in the Kenézlő (KEF) graveyard imply a common substrate of Huns and Conquerors.
@vAsiSTha
Khvalynsk isn't older than steppe_en. The Khvalynsk samples are older than the three steppe_en samples currently available.
But despite this it's easy to see that Khvalynsk got its southern ancestry from a population like steppe_en from north of the Caucasus.
That's because this is the most sensible option, and it fits the data. It's also supported by soon to be published data showing individuals southwest of Samara that are mixtures of EHG and CHG, but without any West Siberian ancestry, and with C14 dates as old or even older than the oldest Khvalynsk samples.
Also, the Khvalynsk population only has a few per cent of West Siberian ancestry, and this is tied to the fact that it was located very close to West Siberia. That's probably also where its Q-M25 comes from.
In fact, Q-M25 is originally from Siberia and it subsequently spread both into far Eastern Europe and Central Asia, and just happens to be most common in Central Asia today. You'll see this reflected in the ancient DNA sooner or later.
At some point you'll have to concede that I know a lot more about this stuff than you do, and I'm a lot better at tying it all together than you ever will be.
Matt (and also Dave):
First of all, let me express my appreciation of your originality, plus technical sophistication, when it comes to exploring the strengths, but also limitations of the modelling tools we have at hand.
Matt, you have pointed out that a comparison of G25-based models and modern populations fares rather poorly in the Volga-Urals area. That area was predominantly Turkic-speaking from at least the early Medieval (Volga-Bulgars, Khazars) to some times between Ivan the Terrible's conquest of Kazan, Catherine the Great's expansions into the Steppe, and Stalin's deportations of Crimean Tartars plus various other groups. So, I think it is fair to assume that modern populations there still bear some Turkic ancestry.
I don't want to turn this into a discussion about the proto-Altaic homeland [ you apparently seem to be interested in this question as much as I am, but we'll find a better time to discuss it in more detail]. Suffice to say for the moment that I think that the ethno-linguistic genesis of Proto-Altaic in all likelyhood occured not too far away from Inner Mongolia, and should have involved some kind of Han-like ancestry. Or, more specifically, some yet unsampled ancestry from the UP/Mesolithic Upper/Middle Yellow River basin that in all likelyhood is also reflected in modern Han Chinese.
As such, I feel that rather than announcing (post-)medieval genetic drift in the Volga-Urals area, your findings may point to the need to introduce a fifth, Han-like component into ancestry modelling for at least this part of Europe.
Since we don't have aDNA from the Upper-Middle Yellow River basin yet (and Devil's Gate is in all likelyhood a rather poor proxy of related ancestry/drift), such a "proto-Altaic" element is of course also so far hardly reflected in G25. This might actually be one of the reasons for the "compression effect" noted by you (another reason could be absence of presumaby Basal-heavy ancestry from the Persian Gulf Oasis).
@davidski said
"Khvalynsk isn't older than steppe_en. The Khvalynsk samples are older than the three steppe_en samples currently available.
But despite this it's easy to see that Khvalynsk got its southern ancestry from a population like steppe_en from north of the Caucasus."
Hahahaha. Always knew you would do anything to validate your biases and with 0 integrity. What evidence do you have to prove that Steppe_en culture is older than khvalynsk culture? List 1 such evidence. the fact is that the steppe_en samples are 400-500 yr younger than khvalynsk samples.
"That's because this is the most sensible option, and it fits the data. It's also supported by soon to be published data showing individuals southwest of Samara that are mixtures of EHG and CHG, but without any West Siberian ancestry, and with C14 dates as old or even older than the oldest Khvalynsk samples."
Too bad the progress_en samples absolutely require WSHG(and Iran_N) as is present in I0434 unlike whatever unpublished samples you are are talking about (i will of course never take your word at face value). It absolutely rejects a simple EHG+CHG source like khvalynsk_en for a wshg richer source like I0434.
Target: RUS_Progress_En_avg
Distance: 2.3041% / 0.02304108
76.0 RUS_Khvalynsk_En_I0434
10.6 GEO_CHG
8.6 TJK_Sarazm_En
4.8 RUS_Darkveti-Meshoko_En
0.0 Anatolia_Barcin_N
0.0 IRN_Ganj_Dareh_N
0.0 RUS_Karelia_HG
0.0 RUS_Khvalynsk_En
0.0 RUS_Samara_HG
0.0 RUS_Sosonivoy_HG
0.0 RUS_Tyumen_HG
0.0 UKR_Globular_Amphora
Target: RUS_Khvalynsk_En_scaled:I0434
Distance: 2.8266% / 0.02826592
64.2 RUS_Khvalynsk_En
14.8 TJK_Sarazm_En
8.6 GEO_CHG
7.4 RUS_Tyumen_HG
5.0 RUS_Karelia_HG
0.0 Anatolia_Barcin_N
0.0 IRN_Ganj_Dareh_N
0.0 RUS_Darkveti-Meshoko_En
0.0 RUS_Samara_HG
0.0 RUS_Sosonivoy_HG
0.0 UKR_Globular_Amphora
0.0 UKR_Meso
0.0 UKR_N
0.0 UKR_N_o
0.0 UKR_Sredny_Stog_En_o4
Either way, even without getting into who is ancestral to whom, the affinity of both progress_en and I0434 to wshg as well as central asian proxy pops is clear.
@vAsiSTha
Good luck with your crackpot theories. Maybe one day you'll be able to write a fiction novel.
Crackpots like you come and go here regularly.
@gamerz_J
"I got further interested in this topic, do you have any good model for EHG?"
EHG is actually not that super difficult to model. We have the usual problem of not having the exact right ancestral populations, but keeping that in mind there are reasonably good proximates. The main part is Paleolithic Siberian "ANE" (as AG3 in the lask of a better option) + Paleolithic European proto-WHG. + CHG-related admixture + when proto-WHG is represented by ancient Italy G25 nMonte takes even some Barin_N. My guess for the latter is that the western side of EHG largely came from the Paleolithic Balkan from a population that intermixed with Anatolians both ways. This admixture is missing in Grotta Continenza, hence Barcin_N comes up.
There are example models.
"sample": "RUS_Sidelkino_HG:Average",
"fit": 6.3536,
"RUS_AfontovaGora3": 64.17,
"ITA_Grotta_Continenza_Meso": 27.5,
"Anatolia_Barcin_N": 5.83,
"GEO_CHG": 2.5,
"sample": "RUS_Samara_HG:Average",
"fit": 7.2325,
"RUS_AfontovaGora3": 65.83,
"ITA_Grotta_Continenza_Meso": 24.17,
"GEO_CHG": 7.5,
"Anatolia_Barcin_N": 2.5,
"sample": "RUS_Karelia_HG:Average",
"fit": 6.717,
"RUS_AfontovaGora3": 70.83,
"ITA_Grotta_Continenza_Meso": 25,
"Anatolia_Barcin_N": 4.17,
"GEO_CHG": 0,
"sample": "WHG:Average",
"fit": 2.4826,
"ITA_Grotta_Continenza_Meso": 86.67,
"Anatolia_Barcin_N": 8.33,
"RUS_AfontovaGora3": 5,
"GEO_CHG": 0,
As you can see even WHG takes some Anatolian ancestry when modelled with Italian HG-s, so when you use the WHG type samples as a references to the western ancestry of EHG, Barcin_N won't comes up like this. WHG is a better reference for the actual source in this regard, but I do not like it, because it also has eastern admixture.
Also the fit is muxh better for WHG that to EHG. My guess is that AG3 is not exactly the best reference for what came into Europe at the end of the Paleolithic.
I have no spell-checker where I am writing this comment now, so please forgive me if I mispelled a few words.
@Vincent
I never claimed that pure ANE came into Europe, if it seemed so it just simplified phrasing in specific context. I might call the Paleolothic Siebrian ancetry of EHG "ANE", but I do not mean it a literal way. ANE is a ghost population, we do not have any direct samples from it.
What I am telling again and again European HG-s do not have East Asian ancestry in the amount that exceeds the amount that is covered by AG3 as a refeference. At the same time the East Siberian ancestry in WSHG significantly exceeds that amount, therefore WSHG as we know it is not an important ancestor.
However AG3 itself had esatern admixture and in all likelity the population that actually moved into Europe had some too. There is no mistery here.
Also as Davidski pointed out a single allele under selection do not need much overall genomewide ancestry to be introduced. However for me this is a side point. For the question wether the known WSHG samples representent a population that contributed significant ancestry to EHG and Yamanaya, it is not relevant to know how much ancient East Asian ancestry the Paleoltithic West Siberians had. They had some, but less than the later WSHG sampples, because apparently eastern admixture incerased with time.
I hope I succesfully clairified things. There is no condradiction between my stand on WSHG ancestry and the East Asian origin of those EDAR alleles.
@Slumber
You haven't bothered keeping up with recently published Eastern Steppe genomic study or East Asian genomic study. I'm not surprised by how confused you are, even with all the new emerging evidence.
ANE samples do not have EDAR alleles, it's SNP data on reich lab site is available. It is coming from actual East Asian source outside ANE as Wang/Lazaridi are suggesting.
Not all EHG carry EDAR allele because both ANE and WHG are diluting East Asian admixture/EDAR allele. We have genomic evidence of admixture between ANE and East Asian taking place in Baikal region, population related to them with high ANE is what headed west. This is archaeologically sound as well because that is also when asian cord ceramics arrives in Baikal region.
WSHG/Tyumen is simply proxy to capture that signal. Using proximate sources and accounting drift improves fit considerably.
RUS_Sidelkino_HG
Fit 2.0100
65.4 UKR_Meso
28.2 RUS_Tyumen_HG
6.4 RUS_AfontovaGora3
RUS_Karelia_HG
Fit 3.1358
58.2 UKR_Meso
22.0 RUS_AfontovaGora3
19.8 RUS_Tyumen_HG
Lazaridi and Wang both suggest EHG needing East Asian source outside ANE. Dzudzuana genomic study will be published sooner or later, we'll see how it will turns out.
I am not sure if you even talking to me actually as you keep attackíng statemens that are the opposites of mines. I did not say ANE had those SNPs. I said the late Paleolithic West Siberians who moved into Europe had it. And they had it because of their East Asian admixture. I say exactly what your sources claim in this regard. You are bringing up a contradiction that does not exist.
The WSHG samples might create a window into the origin of the alleles and it is interseting and all, but this entire EDAR question has hardly any relevance for the other question: wether the late Mezolithic West Siberians, aka the population reprezented by the current WSHG samples are an important ancestor of EHG and Yambaya. They cannot be, because they have too much East Asian ancestry. But again - altough I wrote this just a few hours back, but whatever - this doey not mean that the actual Siberian ancestral populations had none.
@vAsiSTha
This is what TreeMix thinks of your Khvalynsk theory.
https://drive.google.com/open?id=1RtA2CcKI6EJbRfJWe6V9CF-KMcr2E7Z0
Bahaha...
"https://drive.google.com/open?id=1RtA2CcKI6EJbRfJWe6V9CF-KMcr2E7Z0"
Lol idiot i never claimed that the 2 high quality khvalynsk samples had anyhing to do with central asia. theyre just EHG + CHG with no central asian or wshg at all. Theyre also younger than I0434 sample.
Im talking about progress_en & Khvalynsk_o I0434 and the other unpublished khvalynsk Q-M25 samples. This I0434 is selected as source over khvalynsk (ehg+chg) or karelia_hg or samara_hg by progress_en, vonyuchka_en, sredni_stog_o4 as well as steppe_maykop. In that way it is extremely central to whole of 5th and 4th millenium PC steppe.
left pops:
Steppe_Eneolithic
Georgia_Kotias.SG 0.118 +- 0.037
Caucasus_Eneolithic 0.124 +-0.037
Russia_Khvalynsk_EN 0.542 +- 0.018
Sarazm_EN 0.216 +- 0.037
pvalue: 0.41
https://pastebin.com/eU9a5q5t
dont throw your stupid treemixes at me. explain the affinity of these pops to sarazm on nmonte as well. CHG & Sarazm are at distance of 15% using G25, vahaduo etc is not stupid.
@vAsiSTha
You just don't have the ability to fill in the gaps.
That's OK, a lot of people don't, but at least they don't get obsessive compulsive about it online.
You should stop making a fool of yourself though, because I'm sure even you realize that thousands of ancient samples are on the way, and nothing will be left to our imaginations.
@Davidski Samul and others.
Is it possible to detect Finland/Finnish specific genetic drift in the genomes of individuals using G25?
Hey Davidski,
I'm no fan of crackpot theories either, but given that you seem like a fan of the Aryan Migration Theory, I was wondering if you could explain these.
https://ibb.co/g6kMRcf
Why are South Asians as closely related to EHG as they are with Yamna. Doesn't makes sense in terms of a Aryan Migration Theory. IF there was only Iran_N in South Asia before Steppe_MLBA admixture, IE Indians would be closer to Yamna than EHG, because Iran N is much closer to CHG and Yamna than to EHG.
So the above graph disqualifies any modelling of South Asian ancestry as anything like ASI+ Iran_N+Steppe_MLBA (Or Steppe_MBA). It needs to be more like ASI+Iran+EHG (or MA1, AG3 etc)
A similar F4 graph with Ror comparing affinity to Yamna vs EHG.
https://ibb.co/DDM5Hpv
The answer to this question will be given by samples of the Abashevo culture from which Sintashta originated. When you see the Abashevo migration path, you will know that it ran through the entire forest strip of the European part of Russia, where it gained EHG. The population of the Catacomb culture ( descendants of Yamnaya) was mostly exterminated, and the remnants of it were pushed partly to the South Caucasus, partly to Europe, and only partially were assimilated
@Vladimir,
That is not the reason.
CHG reflects the affinity of South Asians to the Caucuses. EHG reflects South Asian affinity to MA1 and AG3 populations.
South Asia has very deep affinity to both groups.
So when we model South Asians, we are modelling ancient relationships, for ASI we can use any modern South Indian, then CHG models the bidirectional geneflow between South Asia and West Asia + Caucuses. EHG is needed to model North vs South drift in Indian populations.
CHG is higher in Western Indian and Pakistani groups, whereas the EHG numbers are consistent across most Northern Indian and Pakistani populations.
This represents two different relationships of North Indian groups, one to ANE and one to the Caucuses Middle East cluster. It has nothing to do with any Indo-Aryan migration.
People talking about an Aryan Migration in terms of aDNA don't understand the concepts like EHG, CHG, Steppe_MLBA etc.
The ancestors of EHG, MA1 or AG3, they most likely came about after drift from a population of Northern Indians (probably very similar to modern IAs).
Northern India/Kalash/Central Asia the is point of dispersal between CHG, ANE, and East Asian. Indian populations look like they've been very stable for a very long time.
mzp1 said...
"People talking about an Aryan Migration in terms of aDNA don't understand the concepts like EHG, CHG, Steppe_MLBA etc."
Don't lie, you're a liar. You're the one who doesn't understand. You don't understand anything about any subject, you only carry one verbal diarrhea. Your delirium can't fool anyone because everyone knows you're a moron who writes only delirium imagining he's a connoisseur, because we all see you are disgraced by not knowing anything.
@Mr. J,
Yes, I think there is some Finnish specific drift in G25 PCA. There's three Finnic pops in G25 PCA: Finnish, East Finnish, Karelian.
East Finnish is most pure Finnish. All Finnic pops share drift with them.
"Finnish" fit as 75% East Finnish, 25% Swedish.
Karelian fit as 65% East Finnish, 25% Russian, 10% Saami
Russian Pinega is mostly Karelian: 61% Karelian, 20% Saami, 20% Russian.
@mzp1
Learn the basics and stop trolling here or you're banned.
Last chance.
@Davidski,
Ok, but can you/someone fill me in on what's wrong with what I said about the affinity of North Indians to EHG vs Yamna as evidence against a steppe migration into India?
I guess I should find another outlet for discussions and ideas, not because I don't like it here, but I don't really want to push these ideas here if your not interested.
@mzp1
You can start with these blog posts and maps:
On the doorstep of india
Some myths die hard
R1a: from Europe to India
And do not come back here until you've read them and understand the basics.
@Slumbery
Thanks quite clear! Trying out the Global 25 I got similar results but worse results because I only used Afontova Gora, WHG and CHG. I suspect the Barcin_N signal is due to Dzuduzana ancestry.
However, could you please elaborate on the following?
"I said the late Paleolithic West Siberians who moved into Europe had it. And they had it because of their East Asian admixture. "
How much East Asian admixture do you think they had? I never saw any estimates of more than 3% East Asian in EHG, so I guess it can't be higher than that but in any case I'd be interested in your thoughts about it. You also mean that this East Asian admixture was not ANE mediated as in the same component that Yana had, right?
@Vincent
"This is archaeologically sound as well because that is also when asian cord ceramics arrives in Baikal region.
WSHG/Tyumen is simply proxy to capture that signal. Using proximate sources and accounting drift improves fit considerably."
WSHG had 20% East Asian ancestry. If Karelia had (about) 20% WSHG ancestry then they had about 3-4% East Asian ancestry. I don't know if even that is seen in Yamnaya (possible that not all EHG samples had this admixture) but given the Yamnaya had about half or less EHG, that is about 1-2% East Asian admixture in Yamnaya.
Once again though, you don't know the genomic profile of the cord ceramic industry people. You also don't know if the diffusion of this technology was mediated by admixture.
gamerz_J
"How much East Asian admixture do you think they had?"
I cannot answer this question. Note that nMonte and its formal "sibling" qpAdm work with existing samples and cannot work with ghost populations.
Also they could have different East Asian ancestries from different time depths, so the ancestry paths can be rather braided river like.
Maybe with qpGraph something could be done.
What I thin is this:
East Asian ancestry in the Late Paleolithic Siberian ancestors of European HG-s: x
East Asian ancestry in AG3: y
East Asian ancestry in Tyumen HG and Sosonivoy HG: z
And: 0 < x ≤ y < z
I focused on the y < z part as an argument against late (Tyumen/Sosonivoy time) Siberian ancestry of EHG and Yamnaya.
@Slumberry
Thanks,but I do have a last question about x East Asian ancestry in late Paleolithic Siberian ancestors, are you referring to ancestry via ANE or something else?
@gamerz_J
I am not going to call them ANE now, because using this simplified handle for them apparently causes misunderstandings. EHG and even more western groups had ancestry from people who lived in Western Siberia in the Epipaleolithic. This population was mainly "ANE", but not purely.
Ancestry "via ANE" is definitely not the right term, because this is the opposite: ANE ancestry via Epipaleolithic Western Siberians (who were mostly ANE, but not a reference for pure ANE, whatever is the latter).
"the most widely accepted Indo-European phylogenies show the Anatolian family as the most basal node."
All approaches with the mentioned result are heavily biased by neglecting the Zipf-distribution of the linguistic data and other shortcomings.
archlingo
"By and large, the PC steppe is still seen by historical linguists and archeologists as the most sensible place to put the PIE homeland." < Having just written a book about The Earliest Wheel Finds, their Archeology and Indo-European Terminology in Time and Space, and Early Migrations around the Caucasus, I cannot follow this sentence. Both linguists and archaeologists remain very reserved regarding any homeland hypothesis - perhaps because fearing to loose own terrain.
archlingo
@archlingo
Both linguists and archaeologists remain very reserved regarding any homeland hypothesis - perhaps because fearing to loose own terrain.
Bullshit.
The typical view is that there are no plausible alternatives to the steppe homeland.
Post a Comment