search this blog
Monday, February 14, 2022
Blond hair is only indirectly associated with Anatolian ancestry in Estonia...duh
In a recent paper about complex traits in Europeans, Marnetto et al. found that blond hair and blue eyes showed a relatively high association with ancient Anatolian ancestry.
This is a somewhat curious finding considering that ancient Anatolians weren't particularly blond haired or blued eyed, and that's probably an understatement.
However, the Europeans that Marnetto et al. based their analysis on were Estonians. And in Estonia ancient Anatolian ancestry peaks in the west and north, probably because this is where Estonians have the most Germanic and Finnish ancestry.
Germanic and Finnish populations are somewhat richer in ancient Anatolian ancestry than Estonians, and, unlike ancient Anatolians, they're often exceptionally blond haired and blue eyed.
So it makes sense that, in Estonia at least, ancient Anatolian ancestry is associated with blond hair and blue eyes, but only indirectly so. The more direct link is between Germanic and Finnish ancestry and blond hair and blue eyes.
I feel that Marnetto et al. should've investigated this, and they also should've made it clear that the associations they found won't necessarily be seen in other European countries.
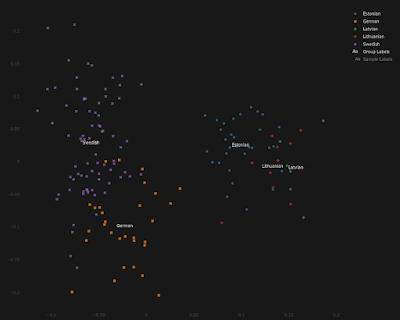
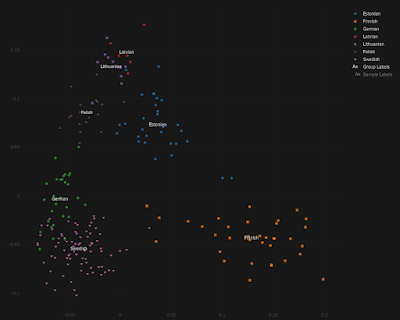
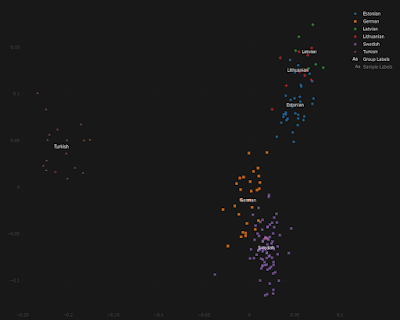
For the doubters out there, and I know there are at least a few of you, below is a series of Principal Component Analyses (PCA) showing how Estonians compare to other populations from around the Baltic Sea, as well as to present-day Turks from central Anatolia.
Note that, by and large, the same Estonians who show more affinity to the Germanic and/or Finnish individuals are also shifted slightly closer to the Turks, and this is because they harbor elevated ancient Anatolian ancestry. The relevant datasheets are available here.
Citation...
Marnetto et al., Ancestral genomic contributions to complex traits in contemporary Europeans, Current Biology (2022), https://doi.org/10.1016/j.cub.2022.01.046
See also...
Ancient ancestry and complex traits in Estonians (Marnetto et al. 2022)
Mainstream media BS: Europeans owe their height to Asian nomads
Subscribe to:
Post Comments (Atom)

228 comments:
«Oldest ‹Older 201 – 228 of 228Yes, according to Lazaridis, Smyadovo I2181 is the oldest M269 we know. Arza in this blog was the first to say that this sample belonged to this lineage and as usual, the Kurganists looked for all kinds of arguments to deny it. However, it is evident that the sample belongs to a farmer of the Gumelnita-Karanovo culture-"Skeleton in flexed position on the left side-Orientated to the East - 110°- The arms were bent, with the palms in front of the face, hips close to the chest and shins were touching the hips. The length of the skeleton in situ is 0.76 m the femur is 0.45 m long. Anthropological determination: male ~25 years old. Grave good included a flint artefact, two ceramic vessels and beads of serpentine, bone and Spondylus"
In other words, R1b-M269 has nothing to do culturally with the steppes. Steppe ancestry?? don't make me laugh, what can you expect if it has been found in an area of contact between both cultures??. Does that prove the origin of M269 in the steppes? Of course NOT- What it proves is that this lineage could have moved in small groups with the neolithic cultures of old Europe and reached Iberia (ATP3, which has an important percentage of Balkan DNA).
*Thank you, we all know the autosomal composition of the Baltic HGs, and we also know that they are P297*, Y13200 and Y13202. We also know that R1b-L754 (Villabruna, Iboussieres)>P297>M269 has never been found in the steppes. R1b is a lineage absolutely linked to WHGs, anyone who understands a little genetics should recognize it immediately.
Regarding the Swiss neolithic farmers Auvernier and Burgaschisee, I would be very careful to accept Furtwängler's data without checking them personally because some of them are clearly wrong.It's very unlikely that any sample with well above 30% Yamnaya has 0%, but I could see some of the Swiss very late Neolithic like Aesch21/MX310/MX304 potentially not even having any when re-checked.
"They've maybe wasted it a little bit on data points formed by using qpAdm in ways that Harney's recent paper would suggest is not the best practice - using pRight that are not the most informative for the populations in question, and which are confounded by a mix of different ascertainment and damage levels"
NO links between L754 and M269?, where do you think M269 came from? from Mars?
Why don't you explain your theory about the origin of R1b-M269?
where do you think this lineage originated?
why do you think it has never been found in the steppes?
Why do you think that mainland Europe is full of R1bs since the Epigravettian?
Why Max Planck has ruled out Yamnaya as the origin of L51>L151>P312?
@Gaska
1) I6222 is phylogenetic consistent and is P310*. What this means is that L23 (the common ancestor of L51 and Z2103) existed at roughly 4100 BCE, right at the time of Srendy Stog and Repin. Afanasievo, Yamnaya, Corded Ware, all share common ancestry, so that means we will see shared clades spread out over many thousands of miles, along with Steppe autosomal DNA. There’s nothing ironic about it. I6222 is a dead end, but he further reinforces the steppe origin of L51. A bunch of Yamnaya belong to the brother clade Z2103, and both Z2103/L51 share a common root in L23; why wouldn’t they come from the same place?
2) I broke it all down for you. PF6518 is found in the following: A00*, C-V20*, C-FT59486, H-Y21612*, I-FT344596, I-FTA37476, (I-BY33169*), I-Y169013, I-FT33727*, I-BY138*, I-Y3712*, (I-CTS10057, I-PH5234), J-MF10501, (J-FGC61612*), L-Y31961*, (T-Y261958, Q-FT380500), Q-Y4943a*, R-Z2123, R-Y88962*, (R1b), (R-M269), (R-FTA35720), (O-MF15397*). Maybe if you read all the other potential calls, you’d see that many other haplogroups can be called for, including J2a-Z6046, R1b-V2219/V88, and I2a-CTS10057. All of them are consistent will the SNPs called by Genetiker.
3) try again. CLL007 has not been carbon dated, and he has steppe autosomal ancestry.
4) cool story bro, and EHU002 has steppe ancestry (62.9% Germany Beaker), so try again
@Gaska
ATP3 tested positive for J2-Z6046-SK2198/S18476. CTS9278/PF5501/M2693, PF6518, and PF6401, are three SNPs in J2-FGC61612*, a subclade of Z6046, while PF2756 corresponds to Z6046 itself. PF2756 is a SNP in I; PF6518 and CTS9278/PF5501/M2693 are SNPs in I-CTS10057; I-Z161*, and I-L801* descend from CTS10057. Genetiker posted his results for ATP3 in September 8, 2015, so at that time, Y16447 was an SNP only found in Y16441. In 2022 however, Y16447 also corresponds to R1b-Z2103>FT31418* (formed 2400 BCE), making it another false positive and remnant of damage. Moreover, the TMRCA of both clades indicates contamination and/or damage. This sample has inconsistent, vague calls, calls which could lead to a multitude of false positives and assignments, such as O-MF15397* and Q1b-FT380500. ATP3 could easily carry a form of I2a, J2a, T1a, or R1b-V2219/V88. Considering the large amount of false SNPs (Y1702, Y16447, Y3545/FGC24163, etc.), PF6401 or PF6518 could be false, meaning ATP3 could belong to G2a-P3147. This sample is that bad; you can work your way to I2a, G2a, T1a, H2, R1b, or J2a. Yet you keep bringing it up, like it’s some smoking gun. We also need his autosomal DNA, because that tells the whole story.
These are contextual samples from roughly the same timespan in Iberia. They indicate what ATP3 could belong to. I lean towards I2a-CTS10057, R1b-V2219/V88, or the EEF related J2a-Z6046>FGC61612*, which the ANF sample I0708 (6224-6074 calBCE) carried. Z6046 has been found in Austrian LBK samples as well.
El Portalón, Atapuerca: ATP5 (Y-DNA: G2a-PF3147, 5214-5015 calBCE), ATP04 (Y-DNA: I2a-M423*, 3336-2937 calBCE), ATP09-443 (Y-DNA: I2a-M436, 3096-2913 calBCE), ATP016/ATP12-1420 (Y-DNA: I2a-Y34539*, 3011-2881 calBCE), ATP17 (Y-DNA: I2a-M223, 3008-2783 calBCE), ATP2 (Y-DNA: H2m-Y21612*, 2900-2675 calBCE), I5838 (Y-DNA: I2a-Y34539*, 2900-2300 BCE)
More contemporaneous Iberian samples: MUR (Y-DNA: G2a-BY133179, 5299-5066 calBCE), I6617 (Y-DNA: I2a-Z161, 2900-2300 BCE), I0406 (Y-DNA: I2a-Z161, 3900-3600 BCE), c40331 (Y-DNA: I2a-Z161, 3765-3636 calBCE), I0455 (Y-DNA: I2a-CTS10057/L623, 2905-2636 calBCE), I0411 (Y-DNA: R1b-V88>Y8451*, 5298-5059 calBCE), I0410 (Y-DNA: R1b-V88>Y8451*, 5298-5057 calBCE), CHA002 (Y-DNA: R1b-V88>Y8451, 5302-5061 calBCE)
Regarding Patterson's English R1b, the only thing you can think to say is that they have percentages of steppe ancestry?
What do you think about the dating and their archaeological contexts?
If the dating is not wrong, we have steppe ancestry in the British Isles 500-1000 years before the arrival of the BBC to the islands. If you have read Patterson's paper carefully you will see that the first P312 BBs in England are High_EEF, that is, a kind of outliers. This means that they can NOT have their origin in the steppes, but in France or Iberia. The whole steppe theory is a permanent contradiction
In summary you have L754 in Italy and France, P297 in the Baltics, Scandinavia and the Balkans, M269 in cultures of old Europe, Western megalithic culture (Iberia, England), L51 in Swiss and Iberian farmers, L151-U106 in Bohemia (Central Europe). And you still defend an origin in the steppes or in Mongolia of these lineages?
Only V88 (originating in the Balkans), V1636 (Khvalynsk, Eneolithic Progress and Yamnaya) and Z2103 (Yamnaya, Afanasievo etc) have been found in the steppes. These R1b lineages are the only ones that at the moment can be linked to the expansion of the Yamnaya culture (and therefore to IE). Now we are going to continue looking for L51>L151 in the forest steppe, time will tell who is right.
@Gaska
1) Every R1b-M269 sample has steppe ancestry. ATP3 is not M269; he could be G2a, J2a, T1a, R1b-V88/V2219, or I2a. He has abysmal coverage (37166 SNPs). I2181 has 46.1% Yamnaya in qpAdm, with a standard deviation of 17.4%. In admixture analysis, he picked up EHG, and a larger Yamnaya_Samara component. R1b-M269 is from Eastern Europe, it comes from EHGs. There’s not one M269 sample without EHG/Steppe admixture.
2) I said that the L754 bearing XN191 has nothing to do with M269 and the Steppe, because he’s only L754. V88 also comes from L754, and all the other contemporaneous HGs/EEFs (from Italy, Iberia, Czechia, and Germany) belong to V2219/V88. These are the samples that make it very likely XN191 belonged to V88: I14169, I14176, I14173, I0559, I1593. The aforementioned all belonged to V88, and are from pre-Corded Ware/Beaker Czechia and Germany. R1b-L754>V2219>V88, not L389>P297>M269, make sense bud?
3) ironic. You appeal to authority with Max Planck (without providing any hard evidence) but when the very paper that published the Swiss farmers says they have steppe ancestry, you think they have to be wrong lol. This is called confirmation bias, and you are a class A example of it. Just a reminder of what I told you earlier: “MX304 has 21.5% Yamnaya_Samara (SD is 14.6%), and MX310 has 30% Yamnaya_Samara (SD is 16.9%) according to the authors of the study. Additionally, MX304 is positive for R1b-P311. Check Figure 2 and Supplementary Data 4.”
4) Villabruna was not positive for M269 lol. You just make shit up dude. Villabruna was negative for FGC21012 (V2219 equivalent), P297-equivalents, and V1636-equivalents. Funny, you think Villabruna being R1b-L754* makes an origin in Western European HGs likely, but you insinuate that L51 would have originated in Afanasievo or Mongolian or something, if we accept I6222 being L52* lol. Do you understand phylogeny, like a little bit? Villabruna has ANE anyway, his line came from Eastern Europe.
it seems to me that in the Smydovo sample there is some contradiction: on the one hand they say it lacks CHG ancestry. On the other they say it has Yamnaya ancestry which seems illogical. There is no Yamnaya ( aka steppe) ancestry without CHG. I2181 seems just a round of the mill Ukraine eneolithic sample. The only important thing is that it probably came from the eastern most part of the Ukraine neolithic geographical and genetical continnuum since it has likely a WHG/ANE ratio lower than the average Ukraine neolithic samples.
Another hint that PIE completely lacked steppe ancestry as also the Dereivka samples clearly show. PIE were just WHG/EHG and EEF. Period.
@Gaska
1) The samples have steppe autosomal DNA. The authors themselves said the dating was contaminated. Dating can be wrong, as in the case of the Alexandria R1b. They don’t have levels of contamination that would throw off analysis, only the dated element does, so via deduction, we know that the dating is at fault: “The use of a consolidant on the teeth may have resulted in contamination affecting the radiocarbon determination.” Radio Carbon dating is not perfect. There weren’t high levels of contamination via its X-chromosome too. Stop engaging in confirmation bias.
2) lying again, dude please just stop. There is one L754* in Italy and that’s Villabruna. He’s a dead end. What French sample? P297 was the lineage of the Samara HG, and all the Baltic HGs with P297>Y13200, have EHG, as does VK531. All the British Beakers have steppe DNA. What Balkan samples belong to P297*? And I want full coverage not low coverage. By that logic P originated in SGC. V88 is also in the Ukrainian HGs, and the Swiss farmers have steppe autosomal DNA. The CWC L151 and U106 guys have large amounts of steppe DNA and Q-Z5902, another link with Afanasievo. Iron Gates have EHG. Every time these lineages show up, there’s ANE/EHG. Try again.
I6222 belongs to P310*/L52*. L52*/L52 comes from L51. L51 formed 4100 BCE, and it has a TMRCA date of 3700 BCE. Z2103 (Yamnaya and late Bohemia CW had both) and L51 descend from L23, which formed 4400 BCE; its TMRCA is 4100 BCE. That’s right towards the end of Srendy Stog, Repin, Progress/Vonyuchka, and Khvalynsk. I6222 is a dead end, but his line’s parent lineage (L51) spread out from the Pontic Caspian and/or East European forest steppes, and into the rest of Europe and Asia, with Afanasievo, Yamnaya, and Corded Ware. Anywhere there’s R1a-M198 and R1b-M269, there is Steppe ancestry/EHG. Just because a sample is found somewhere with a certain lineage, doesn’t mean that lineage originated there, especially when we consider autosomal DNA, formation dates, and the clade’s TMRCA. R1a-YP4132 (formation: 10200 BCE, TMRCA: 4200 BCE) didn’t originate in the Andronovo sample I4773 (1463-1568 BCE), despite the fact that he’s the first sample we’ve found with YP4132.
Iboussieres31-2 (10050-9400 BCE) is positive for K (M9 1x C>G), R (F356 2x T>C), and R1-M173(F211 1xT>G); he has one dubious call for R1b-L754 (PF6271, 1xG->A), however, he’s negative for PH155 (BY14355), V2219 (PF6323), and V88 (PF6292). He has low coverage (80100 SNPs). Regardless, we won’t know what he belongs to. He could be L754*/L389* like Villabruna. P297 formed by 13600 BCE; its TMRCA is 11300 BCE. Villabruna (12200-11800 calBCE) and Iboussieres are late to the party as far as relevance is concerned. They aren’t the forebears of P297 (Villabruna was negative for P297 equivalents). If anything, these dead end L754* and L389* clades represent an early wave of ANE/EHG related admixture from the east. This also has little to do with M269, or that fact that anytime M269 is found, Steppe ancestry/autosomal DNA is also found.
@Simon Stevin-
“It is always fun to discuss these issues with stubborn Kurganists who do not understand or do not want to understand the harsh reality. You said
1-“Mongolia-I6222 is a dead end, but he further reinforces the steppe origin of L51”-Yes, it is a dead line because it is a garbage sample, I have already told you to ask for advice or to read the review procedure for this paper. Ergo WISHFUL THINKING. There is no P310 in Mongolia, nor in Yamnaya, Repin, Sredni Stog, Khvalinsk etc etc etc etc..... Maybe you can explain to us how P310 got from Mongolia to Western Europe, I'm sure you have a wonderful theory.
2- “I broke it all down for you. PF6518 is found in the following”- You are using the same tactic that many people have used for years to deny that ATP3 is R1b-M269. You should know that even Harvard-Reich have recognized that this sample belongs to that lineage then they have simply agreed with us. All your attempts to dirty this sample are a red herring to hide the harsh reality..Sorry mate, you can't keep denying the evidence and lying about the dubious calls of this sample because it has already been officially recognized as belonging to this lineage. Game over, we have M269 in the Iberian Neolithic-The case for ATP3 being R1b1a/2-M269 is as follows:
*GHIJK-Equivalent M3773/CTS12673 [Y-28.66.607 C>T] is the first in the series ATP3's SNPs leading to R1b2a/2-PF6518-This SNP is in the Poznik (2013) & Karmin (2015) "callable" regions, and while it's a C>T transition, it's not an A>C or G>T purine to pyrimidine transversion, which could be the result of DNA damage that deaminates purines.
*K-M9-ATP3-PF5501/M2693/CTS9278 [Y-18.795.682 C>T] is a transition in the Poznik (2013) & Karmin (2015) callable regions of the Y, that is equivalent to K-M9. A C>T pyrimidine transition would not likely be the result of DNA damage. The read doesn't have any other changes or signs of damage.
*R1-M173-ATP3 is positive for the R1 equivalent SNP-M748/YSC0000207 [Y-19.045.552 C>T]-Position (hg19)-19.045.552-Position (hg38)-16.933.672-R1b-M748-YSC0000207-rs7892920-C/T-ISOGG-14-C>T is a transition, and the SNP is in the Poznik (2013) & Karmin (2015) callable Y regions
*R1b1a-L389-P297-ATP3 is derived for the R1b1a-P297 equivalent Y97/FGC46/Z8144 [Y-4,646,572 T>G]-ISOGG has Z8144 as G>T, which is backwards, but Ray Banks' tree has Z8144 as T>G, which is correct.-This SNP, a transversion, is the reverse of what would be expected from DNA damage, which would be a G>T, not T>G-
*R1b1a/2-M269:ATP3-PF6518-Marker PF6518 (Y: 23,099,729 A>C) is a transversion, and it is not a C>T or G>A which could be the result of DNA damage.
3-Try again bro, CLL007 is an Iberian Neolithic farmer officially dated to 2,800 BC
4-“Cool story bro, and EHU002 has steppe ancestry (62.9% Germany Beaker), so try again”-Try again what? I suppose you will agree that this sample is Df27, and it is 50 years prior to the case we have in Narbonne (Occitania)-Both are buried in typical sites of the Iberian BB culture (with grave goods that have only been found in Iberia and southern France)-Have you looked at the mitochondrial lineages of these samples?-Their resemblance to German BBs is due to exogamy. EHU002 is the oldest P312 sample in Europe (2434 BC) after Osterhofen (HapY-R1b-P312>L2) Luca Papac, in his work on Bohemia, has made it clear that P312 has nothing to do with the CWC and that its origin must be in a region between the Rhine and the Atlantic coast. In other words, it is Western as we have always defended against many knuckleheads in anthrogenica who said that its origin is in Yamnaya Ha Ha Ha
5-“R1b-M269 is from Eastern Europe, it comes from EHGs”- M269 is from EHGs???, you must be kidding, do you know any EHG that belongs to that lineage? -You should stop talking nonsense
6-“These are the samples that make it very likely XN191 belonged to V88” That sample is a neolithic farmer R1b-L754 and it is negative for V88, so try to adjust to reality
7-“You appeal to authority with Max Planck (without providing any hard evidence)”-Hard evidence???, Max Planck has recognized what everyone knew (only the stubborn kurganists denied it) that there is no genetic continuity between Yamnaya and the CWC- Sorry mate but I was right and all of you were wrong.
8-I have already explained to you that Furtwängler has mistakenly used qpAdm, those neolithic farmers have close to ZERO steppe ancestry. Why don't you run the samples?
9-“Villabruna was not positive for M269 lol. You just make shit up dude” Who said that Villabruna was M269? It is L754 and if you understand a bit of phylogeny, you will understand that this SNP is upstream of P297 and M269. Have you found any L754 in Ukraine or Russia? NO, then currently that lineage has Western origin, there is nothing to discuss about it.
89,88-WHG/UHG
7,16-ANE
0,52-ASE
0,97-East-Eurasian
0,36-West African
1,12-East-African
10-Phylogenic evidence clearly indicates the common ancestors of extant L51 most recently lived in Western Europe, most likely France-The first subclade to break away from L51 not related to L52 is R1b-Z2118, dates back to only 400 years after the formation of L51 (5700 years ago, so well before the migrational period of L51 Beaker folk across Western and Central Europe). The men with this subclade, in modern times, are distributed mostly around Southern France and the Rhône region. Why is that the case, if not for that general area being L51's homeland? Why, during the great Beaker migrational period, would already differentiated Z2118 men "choose" to migrate to Southern France and not throughout the rest of Western and Central Europe? It would be like time-travelling to just before the great migrational period of the Beaker folk, marking those carriers of the subclade Z2118, and seeing that the vast majority migrated to that region North of the West Med. - that is ridiculously unlikely!-An Eastern European origin of L51 would require those with branches that split at an earlier date before the great migrational period of the Beaker folk (i.e. Z2118) to have preferentially, for some reason, migrated to the vicinity of the South of France, and not elsewhere, DESPITE having been present at the earliest stages in L51's Urheimat. It would be like travelling back in time to just before the supposed great migration of L51 Westwards from E. Europe, marking those who carried this haplogroup, and seeing that the vast majority of them ended up in Southern France and the areas nearby and not so much elsewhere. There IS no reason for that, there can't be!
11-“What French sample?-Iboussieres31-2 (9.775 BC)-France Epipaleolithic-HapY-R1b-L754 (xV88)-mtDNA-U5b2/b-You already have another WHG belonging to this lineage. do you understand?
12-“P297 was the lineage of the Samara HG” Ha Ha Ha
13-“and all the Baltic HGs with P297>Y13200, have EHG, as does VK531”-They are all fundamentally WHGs, I recommend you check their mitochondrial lineages because those are Ukrainian. I repeat R1b is a marker of the WHG nothing to do with EHG.
14-“All the British Beakers have steppe DNA”-The samples we are talking about are neolithic farmers apparently with steppe ancestry, nothing to do with the BBC.
15-“What Balkan samples belong to P297*?-The Balkans and the rest of Europe are plagued by V88 and P297, and the samples are older than those that have appeared in the Ukraine. You get it?, the WHGs colonized the Ukraine and the steppes, not the other way around.
*I4081 (7.385 BC)-Ostrovul Corbului, Iron-Gates-HG-HapY-R1b1a-L754 (xPF6340)-mtDNA-H13
*I4655 (6.815 BC)- Schela Cladovei, Romania-HapY-R1b1a-mtDNA-K1
*I5411 (6.650 BC)- Schela Cladovei, Romania-HapY-R1b1a (x R1b1a/1a2-M269)-mtDNA-U5a1/c1
*I5408 (5.500 BC)-Ostrovul, Corbului, Iron Gates HG-HapY-R1b1a-L754-mtDNA-K1c-
@Gaska
1) let me repeat this again: P310* is at the L52 phylogenetic level. L52*/L52 comes from L51. L51 formed 4100 BCE, and it has a TMRCA date of 3700 BCE. Z2103 (Yamnaya and late Bohemia CW had both) and L51 descend from L23, which formed 4400 BCE; its TMRCA is 4100 BCE. That’s right towards the end of Srendy Stog, Repin, Progress/Vonyuchka, and Khvalynsk. I6222 is a dead end, but his line’s parent lineage (L51) spread out from the Pontic Caspian and/or East European forest steppes, and into the rest of Europe and Asia, with Afanasievo, Yamnaya, and Corded Ware. You what a TMRCA is right? That means all Z2103 and L51 men share a common male ancestor, who lived around 4100 BCE. How did P310* get to Mongolia? Are you serious? Afanasievo spread eastward from the Pontic-Caspian steppe and into Asia, along with Z2103, J1, and Q1b.
2) not an argument. Results are from YFull 2022, the parentheses indicate a possible chain of calls.
PF2756: J2-M102, J2-PF5008, J2-PF5160, J2-BY186473, J2-Y64860, (J2-Z6046), J2-Z6064, J2-CTS4800, J2-Y7687, F-Y27277, J1, (I, K, G, H), and E-M250.
M3773/CTS12673: (F), C-Y138372*, and A-FT174073
CTS9278/PF5501/M2693: (J-FGC61612*, G-PF3177*, K, I-CTS10057, I-BY33169*), I-BY138*, (I-PH5234), and I-BY3605*.
PF6518: A00*, C-V20*, C-FT59486, H-Y21612*, I-FT344596, I-FTA37476, (I-BY33169*), I-Y169013, I-FT33727*, I-BY138*, I-Y3712*, (I-CTS10057, I-PH5234), J-MF10501, (J-FGC61612*), L-Y31961*, (T-Y261958, Q-FT380500), Q-Y4943a*, R-Z2123, R-Y88962*, (R1b), (R-M269), (R-FTA35720), (O-MF15397*)
PF6401: A00*, E-Y175024, E-Y45797, E-L241*, C-V20*, C-FT59486, C-Y10492, C-M208*, C-Y507, C-Y11606*, G-Z36520*, G-FGC34434*, G-PF3177*, G-BY133179, G-Y144865, G-Z6128*, G-BY146155*, G-Z1903*, H-P96*, H-Y19962*, I-FGC21819, I-F1295*, I-FT344596*, I-FT344665*, I-FT344602, I-Y28222*, I-FTA37476, I-BY33164*, (I-BY33169*), I-Y79427*, I-L1498, I-FT27731*, I-A8742*, I-BY37286*, I-BY60222*, I-Y4882*, I-Y16473, I-BY193138*, I-S2555*, I-Y52134, I-Y34539*, I-Y7240, I-BY138*, I-Y43545, I-Y3709*, I-FT380000, I-BY166007*, I-BY168618*, I-Y3712*, (I-Z161*, I-PH5234, I-L801*), I-BY3605*, J-Y159482*, J-FTA1458*, J-MF10501, (J-FGC61612*), J-Z36829*, L-Y6288, T-Y13280*, T-Y63197*, (T-Y261958), S-B255*, Q-Y144860*, (Q-FT380500), Q-Y2700*, Q-F2019*, Q-BZ4911*, Q-BZ2222*, Q-Y12449, Q-FT421589*, Q-YP4547, Q-Y4276, Q-Y4277*, Q-M848, R-M459*, R-BY149647*, R-S23592, R-YP1558, R-YP1558*, R-Y874*, R-Z283, R-Y126934, R-Y2395, R-YP986*, R-Z280, (R1b), (R-P297), (R-FTA35718, R-FTA35720, R-Y127541, R-PF6287*, R-PF6362), R-Y3370*, O-K2*, (O-MF15397*)
ATP based on these calls, could belong to R1b-V2219/V88, I2a-CTS10057, T1a-Y261958, or J2a-Z6046>FGC61612*. He also has calls for the following: A00-AF12, E-M78-L547/PF2166/V4057, J2-Z6046-SK2198/S18476, F-Y1811/FGC2054, GHIJK-M3773/CTS12673, F-PF2756, K-CTS9278/PF5501/M2693, O-F1835, Q-Y1109, Q-YP1618, Q-Y18433, I2-Y16441-Y16447 (formed 2100-1800 BCE), R-Y479/F370/PF6047/M708, R1-M748/YSC0000207, R2-Y3545/FGC24163, R1b-P297-Y97/FGC46, and R1b-L21-Y16874-Y17204 (formed 1500 BCE). This is likely contamination combined with low coverage.
@Gaska
4) CLL007 is not carbon dated, and he has steppe autosomal DNA. Invading males with steppe ancestry took wives with no steppe ancestry, hence the non-steppe mtDNA. EHU002 (2562–2306 cal BCE, 62.9% German Beaker) has steppe ancestry/autosomal DNA. P312 descends from L151, and the oldest L151 samples are in Bohemia Corded Ware (STD002, 2882-2673 calBCE). All M269 males have steppe autosomal DNA, there’s not one who has none.
5) All M269 bearing males have steppe autosomal DNA, and yeah it comes from Eastern Europe. Find me one that doesn’t have that autosomal signature in it.
6) let’s break this down once again. XN191 dates to 5316-5081 calBCE. M269 formed 11300 BCE, TMRCA IS 4400 BCE. XN191 didn’t test negative for anything; the most coverage available was to L754, which could lead to V2219/V88. He didn’t test negative for any of the V2219/V88 equivalents. So yes, on the basis of I14169, I14176, I14173, I0559, and I1593, he very likely belongs to V88. But we won’t know for sure because of his low coverage (20283 SNPs). The data certainly doesn’t suggest M269.
7) not an argument, my quote again: “MX304 has 21.5% Yamnaya_Samara (SD is 14.6%), and MX310 has 30% Yamnaya_Samara (SD is 16.9%) according to the authors of the study. Additionally, MX304 is positive for R1b-P311. Check Figure 2 and Supplementary Data 4.” You have not explained in anyway how he used qpAdm incorrectly. You just simply said “they’re wrong,” which isn’t an argument. Those sample are low coverage too (29434 and 28525 SNPs).
8) find me the quote from Max Planck, either way it’s not an argument. Just saying “they agree with me” even if we take you on face value, is not presenting or refuting evidence. Not an argument
9) Villabruna has ANE. Villabruna is negative for FGC21012 (V2219 equivalent), P297-equivalents, and V1636-equivalents. He may belong to L389*, but that’s dubious. L389 formed 15100 BCE, and its TMRCA is 13600 BCE. P297 formed by 13600 BCE; its TMRCA is 11300 BCE. Villabruna dates to 12200-11800 calBCE. Villabruna is therefor not relevant to P297, since it already existed before him, and he didn’t belong to it. Let me remind you what I said again: “Just because a sample is found somewhere with a certain lineage, doesn’t mean that lineage originated there, especially when we consider autosomal DNA, formation dates, and the clade’s TMRCA. R1a-YP4132 (formation: 10200 BCE, TMRCA: 4200 BCE) didn’t originate in the Andronovo sample I4773 (1463-1568 BCE), despite the fact that he’s the first sample we’ve found with YP4132.” So Villabruna is a dead end with ANE. He is not relevant to P297. Just because he was found in northern Italy with L754*/L389*, doesn’t mean that’s where those lines originated. EHG has a lot of ANE. R1 comes from ANE ultimately, so deductively it makes that this clade originated in the most ANE rich groups in Eastern Europe.
10) fascinating story with no scientific evidence or hard data. Z2118 formed 3700 BCE, as did L52. They both descend from L51, and both Z2103 and L51 descend from L23, which formed in 4400 BCE; its TMRCA is 4100 BCE. That’s right towards the end of Srendy Stog, Repin, Progress/Vonyuchka, and Khvalynsk. In order for L51 to be WHG, Yamnaya would also have to descend from this group, because Z2103 and L51 spawned from L23 (TMRCA 4300 BCE), and yet there is not one pure WHG sample in 4500-3500 BCE, with M269, L23, Z2103, or L51. The burden of evidence is on you. Where ever M269 is found, there’s steppe autosomal DNA.
11) already addressed this: “Iboussieres31-2 (10050-9400 BCE) is positive for K (M9 1x C>G), R (F356 2x T>C), and R1-M173(F211 1xT>G); he has one dubious call for R1b-L754 (PF6271, 1xG->A), however, he’s negative for PH155 (BY14355), V2219 (PF6323), and V88 (PF6292). He has low coverage (80100 SNPs). Regardless, we won’t know what he belongs to. He could be L754*/L389* like Villabruna. P297 formed by 13600 BCE; its TMRCA is 11300 BCE. Villabruna (12200-11800 calBCE) and Iboussieres are late to the party as far as relevance is concerned. They aren’t the forebears of P297 (Villabruna was negative for P297 equivalents). If anything, these dead end L754* and L389* clades represent an early wave of ANE/EHG related admixture from the east. This also has little to do with M269, or that fact that anytime M269 is found, Steppe ancestry/autosomal DNA is also found.”
12) Samara_HG I0124, Samara Oblast, Russia, 5660-5535 calBCE, mtDNA: U5a1d, Y-DNA: R1b-Y13200>Y13202, MathiesonNature2015 (after PMDTools: R1b-Y13872)
13) mitochondrial DNA alone tells you nothing about Y-DNA, autosomal DNA tells the whole genetic picture of an individual. This isn’t 2005 anymore dude. Baltic HGs, SHGs, Iron Gates, and the Ukrainian HGs, all have significant levels of EHG, and they also have R1b, R1a, and Q1b, which are all lineages of ANE origin. That means they came from the east, not the west.
14) British farmers that were had contaminated radiocarbon dating, but not genomic contamination (x-chromosome), Therefor the carbon dating was flawed, not the genome and autosomal DNA. They all have Steppe autosomal DNA: I2611 has 59.9% Steppe autosomal DNA, with a 2.5% margin of error; I3019 has 44.2% Steppe with a 4.2% margin of error; I3035 has 46.5% Steppe with a 2% margin of error.
15) baseless assertion and assumption with no archaeo-genomic evidence backing it. All the Iron Gates samples discerned past L754, belong to V2219 or V88, and they all have significant EHG.
Romania_IronGates_Mesolithic I5408, Ostrovul Corbului, Romania, 7022-6485 calBCE, mtDNA: K1i, Y-DNA: R1b-V2219>Y244183*, GonzalezFortesCurrBiol2017
Romania_IronGates_Mesolithic I5411, Schela Cladovei, Romania, 7000-6300 BCE, mtDNA: U5a1c1, Y-DNA: R1b-V2219>Y244183, MathiesonNature2018
Romania_IronGates_Mesolithic I4081, Ostrovul Corbului, Romania, 7581-7191 calBCE, mtDNA: H13, Y-DNA: R1b-L754, MathiesonNature2018. Negative SNPs: P297, V1636, PF6292. Dubious SNP: PF6323 (V2219)
Romania_Iron_Gates_Mesolthic I4655, Schela Cladovei, Romania, 7059-6571 calBCE, mtDNA: K1, Y-DNA: R1 (low coverage, 59405 SNPs), MathiesonNature2018. Negative SNPs: M269, PF6292 (V88). Dubious SNPs: P297, V1636, PF6323 (V2219).
P297’s TMRCA is 11300 BCE; M269 formed by 11300 BCE (TMRCA 4400 BCE, which coincides with Srendy Stog), so none of these guys are relevant. In fact it’s very likely I4655 and I4081 are V2219, just like their brethren I5411 and I5408.
Just for a frame of reference, here are some of their older contemporaries:
Serbia_IronGates_Mesolithic I5235, Padina, Serbia, 9221-8548 calBCE, mtDNA: U5b2c*, Y-DNA: R1b-V88>Y127541, MathiesonNature2018
Serbia_IronGates_Mesolithic I5240, Padina Serbia, 9140-8570 calBCE, mtDNA: U5a1c3*, Y-DNA: R1b-V88>Y127541, MathiesonNature2018
Serbia_IronGates_Mesolithic I5237, Padina, Serbia, 9300-5800 BCE, mtDNA: U5a2f1, Y-DNA: R1b-V88>Y127541, MathiesonNature2018
Correction for rebuttal #2) ATP based on these calls, could belong to R1b-V2219/V88, I2a-CTS10057, T1a-Y261958, or J2a-Z6046>FGC61612*. He also has calls for the following: A00-AF12, E-M78-L547/PF2166/V4057, J2-Z6046-SK2198/S18476, F-Y1811/FGC2054, O-F1835, Q-Y1109, Q-YP1618, Q-Y18433, R1b-Z2103>FT31418* (formed 2400 BCE)/I2-Y16441-Y16447 (formed 2100-1800 BCE), R-Y479/F370/PF6047/M708, R1-M748/YSC0000207, R2-Y3545/FGC24163, R1b-P297-Y97/FGC46, and R1b-L21-Y16874-Y17204 (formed 1500 BCE). This is likely contamination combined with low coverage— 37166 SNPs/0.031 autosomal coverage. Along with Z=2.3 contamination based on damage/non-damage comparison. P297 could be a part of a chain of false positives leading to FT31418 or Y17204, due to contamination.
Reminder of what I said previously: “ATP3 tested positive for J2-Z6046-SK2198/S18476. CTS9278/PF5501/M2693, PF6518, and PF6401, are three SNPs in J2-FGC61612*, a subclade of Z6046, while PF2756 corresponds to Z6046 itself. PF2756 is a SNP in I; PF6518 and CTS9278/PF5501/M2693 are SNPs in I-CTS10057; I-Z161*, and I-L801* descend from CTS10057…This sample has inconsistent, vague calls, calls which could lead to a multitude of false positives and assignments, such as O-MF15397* and Q1b-FT380500. ATP3 could easily carry a form of I2a, J2a, T1a, or R1b-V2219/V88. Considering the large amount of false SNPs (Y1702, Y16447, Y3545/FGC24163, etc.), PF6401 or PF6518 could be false, meaning ATP3 could belong to G2a-P3147. This sample is that bad; you can work your way to I2a, G2a, T1a, H2, R1b, or J2a.”
1-You can repeat it as many times as you want, and the more you do it, the more ridiculous your argument will be. That sample is BS, get it, stop fooling yourself (and the few who read what you write) with stupid arguments. The origin of L51 is not in the steppes but in central Europe, and to date, you have not found that lineage in the steppes or the forest steppe- I don't think you are able to understand it, the rest is wishful thinking.
2-ATP3-is M269, if you don't agree, talk to Harvard, they don't think the same as you, you are making a fool of yourself.
3-CLL007 is R1b-L51-Iberian farmer. There is nothing more to say
1-You can repeat it as many times as you want, and the more you do it, the more ridiculous your argument will be. That sample is BS, get it, stop fooling yourself (and the few who read what you write) with stupid arguments. The origin of L51 is not in the steppes but in central Europe, and to date, you have not found that lineage in the steppes or the forest steppe- I don't think you are able to understand it, the rest is wishful thinking. You should talk to Max Planck and explain to them that you have discovered P310 in Afanasievo and that you have therefore solved the problem of the origin of this lineage. Don't you realize that they are cheating you?
2-ATP3-is M29, if you don't agree, talk to Harvard, they don't think the same as you ergo you are making a fool of yourself.
3-CLL007 is R1b-L51-Iberian farmer. There is nothing more to say
4-There is not even one M269 in the steppes, you understand?, there is only one in the Gumelnita-Karanovo culture. Autosomal DNA?, don't make me laugh, it's a loser argument. You know and understand how autosomal components are transmitted, don't you? There are dozens of samples with autosomal ancestry in Europe that are not R1b, you understand that, don't you?
5) XN191 is L754-There is nothing to discuss about it, everything else is absurd speculation.
@Gaska
According to Blagoje Govedarica and his archaeological data, the first burial mounds / kurgans were established east of the Carpathian Arch in the cultures like Cucuteni, Varna, Suvorovo, etc., see:
(…) III.2. Phase Suvorovo
Stone sceptres and presumably the first burial mounds as signs of power and high rank appear after that in the second, Suvorovo phase of the sceptre bearers (Cucuteni A4; Varna 3, 4300—4200 B. C., Fig. 5: 61, 64—70; 6; 7). These new “wealthy”persons, that is, this burial custom, spread rapidly into the vast steppes, first as far as the area of the Dnieper River (Krivoj Rog, Čapli) and during the Suvorovo phase on to the Caspian Sea and the North Caucasus. (…)
Conflict or Coexistence: Steppe and Agricultural Societies in the Early Copper Age of the Northwest Black Sea Area
Blagoje Govedarica
Could you comment on that, please?
6) You have not analyzed those samples, they are Swiss farmers and they have hardly any steppe ancestry. If you are curious you can check it by yourself (and if you don't know, ask for advice to someone who knows how to handle qpAdm) or any other autosomal tool.
7)-The quote from Max Planck? It is clear that you have not read the paper on Bohemia. “We observe a closer phylogenetic relationship between the Y chromosome lineages found in early CW and BB than in either late CW or Yamnaya and BB. R1b-L151 is the most common Y-lineage among early CW males (6 of 11, 55%) and one branch ancestral to R1b-P312, the dominant Y-lineage in BB. Although it is not possible to determine whether the P312 mutation(s) occurred in one of the early CW R1b-L151 males from Bohemia, we note that most Bohemian BB males are further derived at R1b-L2/S116 (R1b1a1a2b1), in contrast to BB males from England, several of whom are derived at R1b-L21 (R1b1a1a2c1), showing that English and Bohemian BB males cannot be descendants of one another, but rather diversified in parallel. A scenario of R1b-P312 originating somewhere between Bohemia and England, possibly in the vicinity of the Rhine, followed by an expansion northwest and east is compatible with our current understanding of the phylogeography of ancient R1b-L151–derived lineages”
8-Villabruna is L754 and this is the father of P297 and the grandfather of M296 - you understand?, there is nothing like it on the steppes.
9-The burden of evidence is on all those Kurganists who have been telling us a fairy tale about the origin of our lineage for 7 years. There is no M269 or L51>L151 in the steppes - This is the genetic evidence to date -L754 (Italy), P297 and V88 (Baltic, France and the Balkans), M269 (Bulgaria and Iberia) L51 (Switzerland), L151 (Bohemia). The steppes???? Ha Ha Ha, There is nothing more to say.
10-Samara I1024 is what????? You said M269 is a lineage linked to the EHGs, are you kidding me? You should provide evidence to stop making a fool of yourself.
11-Significant levels of EHG? You never cease to amaze me
12-Baseless assertion and assumption with no archaeo-genomic evidence backing it. All the Iron Gates samples discerned past L754, belong to V2219 or V88- Ha Ha Ha, arguing with you is fun
After the failure of the Yamnaya-Afanasievo culture, the Kurganists have to keep looking for evidence to confirm their theory (because it is their right to do so). Of course you can believe what you want, but I remind you what Haak, Mathieson, Lazaridis, Reich, Harvard said in 2015-"A massive migration from the steppes brought IE to mainland Europe thanks to the Yamnaya culture migrations (and to the R1a-M417 and R1b-L51 lineages)-7 years later we are facing the biggest scientific failure in the history of genetics. There is NO L51 or M417 in the steppe cultures. At that time not even Villabruna existed and when Fu published that sample he said textually that he was surprised to find R1b-L754 in Western Europe during the Epigravettian, they simply made a mistake and rushed their conclusions.
R1b-M269 in the steppes???? No way, it's the biggest nonsense ever!
I have always defended the same ideas
1-M269>L51>L151 may have originated in Scandinavia, the Baltic or even the Balkans. It has nothing to do with the steppes and is certainly not linked to IE expansion.
2-The BB culture never spoke an IE language
3-P312 is western - Germany, Switzerland or the French-Cantabrian mountain range.
4-DF27 is Iberian
I have also said hundreds of times that all you have to do is to discover M269>L51>L151 in the steppes or in the forest steppe. To date, have you done it? NO, so your theory at the moment is a fairy tale for naive people like you. To say that L51 has to be in the steppes because you have found Z2103 there is simply imbecility (no wonder because most people's analytical skills are a joke).
@Gaska
1) no he’s not M269. He has numerous other SNP calls, he’s low coverage (37166 SNPs/0.031 autosomal coverage), and he has some contamination (Z=2.3 contamination). Harvard also thought Alexandria was from the Copper Age too. Keep using selective appeals to authority, they aren’t arguments. Parenthesis indicate calls that are SNP consistent. The only SNP linking him to M269, is PF6518. Though it’s nice you conceded considering you offered no sound rebuttal, as usual.
PF6518 applies to: A00*, C-V20*, C-FT59486, H-Y21612*, I-FT344596, I-FTA37476, (I-BY33169*), I-Y169013, I-FT33727*, I-BY138*, I-Y3712*, (I-CTS10057, I-PH5234), J-MF10501, (J-FGC61612*), L-Y31961*, (T-Y261958, Q-FT380500), Q-Y4943a*, R-Z2123, R-Y88962*, (R1b), (R-M269), (R-FTA35720), (O-MF15397*)
PF2756 applies to: J2-M102, J2-PF5008, J2-PF5160, J2-BY186473, J2-Y64860, (J2-Z6046), J2-Z6064, J2-CTS4800, J2-Y7687, F-Y27277, J1, (I, K, G, H), and E-M250.
M3773/CTS12673 applies to: (F), C-Y138372*, and A-FT174073
CTS9278/PF5501/M2693 applies to: (J-FGC61612*, G-PF3177*, K, I-CTS10057, I-BY33169*), I-BY138*, (I-PH5234), and I-BY3605*.
PF6401 applies to: A00*, E-Y175024, E-Y45797, E-L241*, C-V20*, C-FT59486, C-Y10492, C-M208*, C-Y507, C-Y11606*, G-Z36520*, G-FGC34434*, G-PF3177*, G-BY133179, G-Y144865, G-Z6128*, G-BY146155*, G-Z1903*, H-P96*, H-Y19962*, I-FGC21819, I-F1295*, I-FT344596*, I-FT344665*, I-FT344602, I-Y28222*, I-FTA37476, I-BY33164*, (I-BY33169*), I-Y79427*, I-L1498, I-FT27731*, I-A8742*, I-BY37286*, I-BY60222*, I-Y4882*, I-Y16473, I-BY193138*, I-S2555*, I-Y52134, I-Y34539*, I-Y7240, I-BY138*, I-Y43545, I-Y3709*, I-FT380000, I-BY166007*, I-BY168618*, I-Y3712*, (I-Z161*, I-PH5234, I-L801*), I-BY3605*, J-Y159482*, J-FTA1458*, J-MF10501, (J-FGC61612*), J-Z36829*, L-Y6288, T-Y13280*, T-Y63197*, (T-Y261958), S-B255*, Q-Y144860*, (Q-FT380500), Q-Y2700*, Q-F2019*, Q-BZ4911*, Q-BZ2222*, Q-Y12449, Q-FT421589*, Q-YP4547, Q-Y4276, Q-Y4277*, Q-M848, R-M459*, R-BY149647*, R-S23592, R-YP1558, R-YP1558*, R-Y874*, R-Z283, R-Y126934, R-Y2395, R-YP986*, R-Z280, (R1b), (R-P297), (R-FTA35718, R-FTA35720, R-Y127541, R-PF6287*, R-PF6362), R-Y3370*, O-K2*, (O-MF15397*)
Other SNPs from ATP3: A00-AF12, E-M78-L547/PF2166/V4057, J2-Z6046-SK2198/S18476, F-Y1811/FGC2054, O-F1835, Q-Y1109, Q-YP1618, Q-Y18433, R1b-Z2103>FT31418* (formed 2400 BCE)/I2-Y16441-Y16447 (formed 2100-1800 BCE), R-Y479/F370/PF6047/M708, R1-M748/YSC0000207, R2-Y3545/FGC24163, R1b-P297-Y97/FGC46, and R1b-L21-Y16874-Y17204 (formed 1500 BCE)
Here are chains of consistent calls that can be made for ATP3 (which assumes all these SNPs are legitimate in the first place):
1. F-Y1811/FGC2054-M3773/CTS12673>I-PF2756>I-CTS10057-CTS9278/PF5501/M2693-PF6518>I-Z161*-PF6401
2. F-Y1811/FGC2054-M3773/CTS12673>I-PF2756>I-CTS10057-CTS9278/PF5501/M2693-PF6518>I-L801*-PF6401
3. F-Y1811/FGC2054-M3773/CTS12673>J-Z6046-SK2198/S18476-PF2756>FGC61612*-PF6401-CTS9278/PF5501/M2693-PF6518
4. F-Y1811/FGC2054-M3773/CTS12673>K-CTS9278/PF5501/M2693-PF2756>R-Y479/F370/PF6047/M708>R1-M748/YSC0000207>R1b-PF6401>R1b-V2219>FTA35720-PF6518
5. F-Y1811/FGC2054-M3773/CTS12673>K-CTS9278/PF5501/M2693-PF2756>R-Y479/F370/PF6047/M708>R1-M748/YSC0000207>R1b-PF6401-PF6518
6. F-Y1811/FGC2054-M3773/CTS12673>K-CTS9278/PF5501/M2693-PF2756>R-Y479/F370/PF6047/M708>R1-M748/YSC0000207>R1b-PF6401>R1b-PF6518>R1b-V88>Y127541-PF6401
7. F-Y1811/FGC2054-M3773/CTS12673>K-CTS9278/PF5501/M2693-PF2756>R-Y479/F370/PF6047/M708>R1-M748/YSC0000207>R1b-PF6401>R1b-PF6518>R1b-V88>PF6287*-PF6401
8. F-Y1811/FGC2054-M3773/CTS12673>K-CTS9278/PF5501/M2693-PF2756>R-Y479/F370/PF6047/M708>R1-M748/YSC0000207>R1b-PF6401>R1b-PF6518>R1b-V88>PF6362-PF6401
9. F-Y1811/FGC2054-M3773/CTS12673>K-CTS9278/PF5501/M2693-PF2756>R-Y479/F370/PF6047/M708>R1-M748/YSC0000207>R1b-PF6401>R1b-V2219>FTA35718-PF6518
10. F-Y1811/FGC2054-M3773/CTS12673>K-CTS9278/PF5501/M2693-PF2756>T-Y261958-PF6518-PF6401
@Gaska
These are not the only possible calls, but they are the more likely ones. PF6287* has been found in Brazil, and in remains from Croatia (sample ID: I26735, 1 CE-100 BCE), while PF6362 has been found in Sardinia (subclade M18), and the Ukrainian HG I1734 (7451-7056 calBCE). It’s likely then that ATP3 (3516-3365 calBCE) belongs to V2219/V88 or I2a-CTS10057.
Confirming this are contemporary samples from El Portalón, Atapuerca: ATP5 (Y-DNA: G2a-PF3147, 5214-5015 calBCE), ATP04 (Y-DNA: I2a-M423*, 3336-2937 calBCE), ATP09-443 (Y-DNA: I2a-M436, 3096-2913 calBCE), ATP016/ATP12-1420 (Y-DNA: I2a-Y34539*, 3011-2881 calBCE), ATP17 (Y-DNA: I2a-M223, 3008-2783 calBCE), ATP2 (Y-DNA: H2m-Y21612*, 2900-2675 calBCE), I5838 (Y-DNA: I2a-Y34539*, 2900-2300 BCE)
and more contemporaneous Iberian samples: MUR (Y-DNA: G2a-BY133179, 5299-5066 calBCE), I6617 (Y-DNA: I2a-Z161, 2900-2300 BCE), I0406 (Y-DNA: I2a-Z161, 3900-3600 BCE), c40331 (Y-DNA: I2a-Z161, 3765-3636 calBCE), I0455 (Y-DNA: I2a-CTS10057/L623, 2905-2636 calBCE), I0411 (Y-DNA: R1b-V88>Y8451*, 5298-5059 calBCE), I0410 (Y-DNA: R1b-V88>Y8451*, 5298-5057 calBCE), CHA002 (Y-DNA: R1b-V88>Y8451, 5302-5061 calBCE).
2) Awesome, that’s not a refutation of any of my arguments. All you could piece together was “ugh you’re wrong.” L51 formed 4100 BCE, and it has a TMRCA date of 3700 BCE. Z2103 and L51 descend from L23, which formed 4400 BCE; its TMRCA is 4100 BCE. That means according to your baseless theories, that the Z2103 in Yamnaya also has to come WHG, and that L23 is a WHG marker. Unfortunately there is not one WHG sample with L23, L51, Z2103, or M269. Anytime those clades are found, there is Steppe/EHG autosomal DNA. Still waiting for those pure, 4500-3900 BCE, WHG, M269 samples from you. You won’t find any.
3) CLL007 has steppe autosomal DNA, a steppe related Y-DNA, and he’s not been carbon dated. From the paper: “CLL007 (Arc. ID: Lech 11 (LE11B)): Adult male burial. This individual was considered intrusive and thus excluded from the analysis, as it showed steppe-related ancestry.” Yup you have nothing to say, destroys your whole point
4) I could not understand your point (you wrote with very bad grammar). Seems like you don’t understand autosomal DNA. That’s the non-sex chromosomes, the other 22 pairs (44 all together), the DNA we get from all of our ancestors. It tells the whole genealogical story, unlike a single chromosome you get from either parent. Autosomal DNA is literally more genomic data (more coverage). Autosomal DNA and full genomic coverage defeat your theories centered around an indigenous, WHG paternal origin for Basques, which is why you don’t acknowledge it. There’s a reason no one here takes you seriously. You argue in bad faith, you have confirmation bias, and you’re nationalist ideologue zealot who cherry picks incomplete, low quality data to suit his world view, instead of honestly looking at the whole picture of available evidences.
5) glad we agree, XN191 has very low coverage (20283 SNPs), he can’t be discerned past L754, and all his contemporaries (I14169, I14176, I14173, I0559, and I1593) belong to V88. He has nothing to do with M269.
@Gaska
6) that’s another non-argument from you. You are saying they don’t have steppe, they paper said they do. Here’s the results of the paper again via Figure 2 and Supplementary Data 4: MX304 has 21.5% Yamnaya_Samara (SD is 14.6%), MX310 has 30% Yamnaya_Samara (SD is 16.9%). Both samples have a low number of SNPs (29434 and 28525 respectively. Either provide evidence as to why they don’t have steppe autosomal DNA (like hard data such as D/F-stats, apAdm, admixture, etc.), or stop wasting my time with your “no they don’t have steppe, but I won’t prove it” responses.
7) Did you even read the paper? Max Planck argued that Corded Ware was contemporaneous to Yamnaya, and that neither derives paternally from the other, but they share a common steppe ancestry; this is likely Sredny Stog or Repin—going by the TMRCA of L23 (4100 BCE). Corded Ware was the East European forest steppe side of the migration, Yamnaya was the Balkans, the Carpathian Basin, and the Pontic-Caspian steppe side of the migration; Afanasievo was the Central/North Asian side. From the paper: “CW individuals were shown to be genetically distinct from culturally pre-CW people, having ~75% of their ancestry similar to Yamnaya individuals from the Pontic-Caspian steppe (3, 4, 23–27). This Yamnaya-like “steppe” ancestry then spread rapidly throughout Europe, reaching Britain, Ireland, the Iberian Peninsula, the Balearic Islands, Sardinia, and Sicily before the end of the third millennium BCE (5, 28–32)…Therefore, despite their sharing of steppe ancestry (3, 4) and substantial chronological overlap (45), it is currently not possible to directly link Yamnaya, CW, and BB groups as paternal genealogical sources for one another, particularly noteworthy in light of steppe ancestry’s suggested male-driven spread (23, 41–43) and the proposed patrilocal/patriarchal social kinship systems of these three societies (46–48)…the addition of either Latvia_MN (57 of 95 supported models), Ukraine_Neolithic (53 of 95 supported models), or PittedWare (32 of 95 supported models) to the sources drastically increases the number of supported models (table S17). These results show the presence of excess Latvia_MN/Ukraine_Neolithic/PittedWare-like ancestry in Bohemia_CW_Early relative to all known Yamnaya and central European Neolithic groups. Our models suggest that this ancestry accounts for ~5 to 15% of the Bohemia_CW_Early gene pool (table S17).” So they share a common Eastern European, steppe related source population (CHG + EHG) via autosomal DNA, and L51 and Z2103 have a common root in L23, but Yamnaya and CWC don’t directly, paternally derive from one another. Yamnaya has 10-20% WHG/EEF, while Bohemia CW has 5-15% WHG/EHG ancestry, on top of their 75% Yamnaya-related autosomal DNA. They all have steppe ancestry still; Max Planck just added even more evidence to the fact that M269 is linked to Steppe peoples/Steppe autosomal DNA. What’s your point? This paper completely shreds your L51=WHG nonsense. Once gain, there was not one M269 bearing, pre-Steppe individual. Your same logic could apply to R1a. R1a has never been found in Yamnaya, but it has been found in CWC, so therefor it’s WHG lol. You think L51 is WHG because CWC has 5-15% EHG/WHG (their WHG is even less because EHG heavy groups like CCC are baked in there too), but so does Yamnaya (10-20%). Is Q1b-Z5902 WHG too? It was found in both Bohemia CW and Afanasievo, and the latter has about as much WHG as Yamnaya_Samara. Both Bohemian CW (the sample OHR001) and Yamnaya have Z2103, is Z2103 WHG now? It has to be if L51 is, because L51 shares a root with Z2103 in L23, and the latter has a TMRCA of 4100 BCE. Where are the pure WHGs from France with R1a, M269, and L23 Gaska? Oh that’s right they don’t exist. You’ll be silenced once those L23/L51 Sredny Stog guys get published.
@ Gaska
8) going over the same stuff again and again is a theme with zealots. Villabruna is not the paternal ancestor of P297. Here’s my earlier rebuke again. Villabruna has ANE autosomal DNA. Villabruna is negative for FGC21012 (V2219 equivalent), P297-equivalents, and V1636-equivalents. He may belong to L389*, but that’s a dubious SNP. L389 formed in 15100 BCE, and its TMRCA is 13600 BCE. P297 formed by 13600 BCE; its TMRCA is 11300 BCE. M269 formed in 11300 BCE. Villabruna dates to 12200-11800 calBCE. Villabruna is therefor not relevant to P297, since P297 already existed before him, and he was negative for P297 equivalents. Just because a sample is found somewhere with a certain lineage, doesn’t mean that lineage originated there, especially when we consider autosomal DNA, formation dates, subclades/SNPs, and the clade’s TMRCA and formation date. R1a-YP4132 (formation: 10200 BCE, TMRCA: 4200 BCE) didn’t originate in the Andronovo sample I4773 (1463-1568 BCE), despite the fact that he’s the first and only ancient sample we’ve found with YP4132. So Villabruna is a dead end with ANE. He is not relevant to P297—he’s negative for it. Just because he was found in northern Italy with L754*/L389*, doesn’t mean that’s where those lines originated. EHG has a lot of ANE. R1 comes from ANE ultimately, so deductively it makes sense that this clade originated in the most ANE rich groups in Eastern Europe. L754’s TMRCA is 15100 BCE; L389’s TMRCA is 13600 BCE. That’s plenty of time (about 1500-3300 years before Villabruna) for dead end rouge clades like L389*/L754*/V88 to travel from Eastern Europe, to northern Italy and France. Villabruna is possibly L389*, while Iboussieres31-2 has one dubious call for L754. One can’t be discerned further, and the other is a dead end with ANE. Neither tells us the origins of P297, L389, L754, or M269. L754 and L389 formed 18400 BCE and 15100 BCE respectively; that’s way before Villabruna, like 3000-9300 years before him. Clades can go places in that stretch of time, and die off.
9) I6222 is L51>L52*/P310*, and no M269 males have been found without steppe admixture. You still can’t find a single one. Until you find a M269 male without steppe autosomal DNA, you are just screaming sweet nothings into the wind, like a zealot shouting about end times.
10) https://docs.google.com/file/d/1CsOR69HpiEKkLjkavAtcOXx17SPUdLfm/edit?usp=docslist_api&filetype=msexcel. Calls for I0124 were done by Sergey Malyshev, with data from Wang et al. (2018). I0124 is R1b-P297>Y13200>Y13202-Y13872
11) yeah all Iron Gates have EHG autosomal DNA, are you new to this? I previously typed an essay for you filled with references about the EHG autosomal DNA in Baltic HGs, SHGs, and VK531. From Mathieson et al. (2018): “Modeling Iron Gates hunter-gatherers as a mixture of WHG and EHG (Supplementary Table 3) shows that they are intermediate between WHG (~85%) and EHG (~15%). However, this qpAdm model does not fit well (p=0.0003, Supplementary Table 3) and the Iron Gates hunter-gatherers show an affinity towards Anatolian Neolithic, relative to WHG (Supplementary Table 2). In addition, Iron Gates hunter-gatherers carry mitochondrial haplogroup K1 (7/40) as well as other subclades of haplogroups U (32/40) and H (1/40) in contrast to WHG, EHG and Scandinavian hunter-gatherers who almost all carry haplogroups U5 or U2. One interpretation is that the Iron Gates hunter-gatherers have ancestry that is not present in either WHG or EHG. Possible scenarios include genetic contact between the ancestors of the Iron Gates population and a NW Anatolian-Neolithic-related population, or that the Iron Gates population is related to the source population from which the WHG split during a re-expansion into Europe from the Southeast after the Last Glacial Maximum.17,37.” Sorry they have EHG too; that’s where their R1b lines came from. Another swing and a miss for you
@ Gaska
12) this really hasn’t been an argument. I’ve presented evidence, thoroughly deconstructed and debunked your fallacious arguments, and you’ve retorted back sarcastically with no evidence or rebuttals, just “no that’s wrong.” That’s 3 year old stuff. You have less evidence to back anything you’ve said than I do. All majority WHG groups with R1b, have V88, not M269 or even P297. They don’t even have that much R1, they are mostly I. R is a minor lineage in WHG heavy populations. Anywhere there is M269, there is steppe autosomal DNA. Anytime we have ancient, full coverage remains with R, they have ANE related DNA. Sorry buddy this isn’t 2004, the days of relying on low coverage mtDNA and Y-DNA from the present day are over. I know it was cute while it lasted, must have felt great to think Basques were the unaltered descendants of Cro-Magnon. Oh well, they’re not. Your ideas are backed by nothing but your own ethnocentric bias. All Iron Gates HGs belong to V2219/V88 (the ones that have coverage past L754 do anyway). Not a single one has calls for L389 or P297. You presented no rebuttal in that regard. All Iron Gates HGs have around 15% EHG. Anyway, thanks for conceding, and for your non-arguments. The oldest M269 is I2181; he has steppe/EHG autosomal DNA (46.1% Yamnaya in qpAdm, with a standard deviation of 17.4%), and he’s on a cline towards Khvalynsk (along with the Varna outlier), unlike the other Chalcolthic Balkan samples. I’ll say again, what M269 sample has no EHG and Steppe autosomal DNA? Until you come back with a sample and an answer, you have no arguments.
@Davidski
Wasn’t this guy banned for his nonsense, Basque nationalist cope fairy tales? How did he get back on the blog?
@Simon
You won't change his mind, no matter what, so there's no point.
Language has one root, but genes have multiple roots.
Post a Comment