When used properly, Principal Component Analysis (PCA) is an extraordinarily powerful tool and one of the best ways to study fine-scale genetic substructures within Europe.
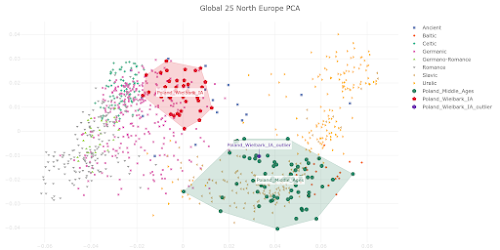
The PCA plot below is based on Global25 data and focuses on the genetic relationship between Wielbark Goths and Medieval Poles, including from the Viking Age, in the context of present-day European genetic variation.
I'd say that it's a wonderfully self-explanatory plot, but here are some key observations:
- the Wielbark Goths (Poland_Wielbark_IA) and Medieval Poles (Poland_Middle_Ages) are two distinct populations
- moreover, the Wielbark Goths form a relatively compact Scandinavian-related cluster and must surely represent a homogenous population overwhelmingly of Scandinavian origin
- on the other hand, the Medieval Poles form a more extensive and heterogeneous cluster that overlaps with present-day groups all the way from Central Europe to the East Baltic, and that's because they are likely to be in large part of mixed origin
- I know for a fact that at least some of these early Poles harbor recent admixture, because their burials are similar to those of Vikings and their haplotypes have been shown to be partly of Scandinavian origin (see here)
- one of the Wielbark females is an obvious genetic outlier (Poland_Wielbark_IA_outlier), and basically looks like a first generation mixture between a Goth and a Balt.
Please note that the PCA is only based on relatively high quality genomes, so as not to confuse the picture with spurious results and noise. Also, all outliers with potentially significant ancestry from outside of Central, Eastern and Northern Europe were removed from the analysis. The relevant datasheet is available
here.
However, sanity checks are always important when studying complex topics like fine-scale genetic ancestry. To that end I've prepared a graph based on f3-statistics of the form f3(X,Cameroon_SMA,Estonia_BA)/(X,Cameroon_SMA,Ireland_Megalithic), that reproduces the key features of my PCA. The relevant datasheet is available
here.
Polish groups from the Middle Ages are marked with the MA suffix, while the Iron Age Wielbark Goths are marked with the IA suffix.
If you're wondering why I plotted the f3-statistics that I did, take a look at this (all groups largely of Scandinavian origin are emboldened):
f3(X,Estonia_BA,Cameroon_SMA)
Poland_Legowo_MA 0.226406
Poland_Ostrow_Lednicki_MA 0.225996
Poland_Plonsk_MA 0.225017
Poland_Trzciniec_Culture 0.224215
Poland_Lad_MA 0.224142
Poland_Viking 0.223838
Poland_Niemcza_MA 0.223659
Poland_Weklice_IA 0.223549
Poland_Kowalewko_IA 0.222584
Poland_Pruszcz_Gdanski_IA 0.222324
Sweden_Viking 0.222091
Russia_Viking 0.222042
Poland_Maslomecz_IA 0.221914
Norway_Viking 0.221825
Denmark_EarlyViking 0.221257
Denmark_Viking 0.221174
England_Viking 0.220979
f3(X,Ireland_Megalithic,Cameroon_SMA)
Poland_Maslomecz_IA 0.219816
Poland_Weklice_IA 0.219501
Denmark_Viking 0.2192
Poland_Kowalewko_IA 0.219176
Poland_Ostrow_Lednicki_MA 0.218916
Norway_Viking 0.218854
Poland_Pruszcz_Gdanski_IA 0.218684
Sweden_Viking 0.218626
Denmark_EarlyViking 0.218529
England_Viking 0.218308
Russia_Viking 0.217999
Poland_Viking 0.217914
Poland_Plonsk_MA 0.217756
Poland_Lad_MA 0.217719
Poland_Legowo_MA 0.21765
Poland_Niemcza_MA 0.217001
Poland_Trzciniec_Culture 0.216551
Interestingly, the Middle Bronze Age samples associated with the Trzciniec Culture (Poland_Trzciniec_Culture) show a closer genetic relationship to Medieval Poles than to Wielbark Goths or Northwestern Europeans. This is indeed the case both in terms of genome-wide and uniparental markers, including some very specific lineages under Y-chromosome haplogroup R1a.
But that's a much more complex issue that I'll leave for another time. So please stay tuned.
See also...
Slavs have little, if any, Scytho-Sarmatian ancestry

261 comments:
1 – 200 of 261 Newer› Newest»The Wielbark outlier looks like a 50/50 Goth/Balt mix and clusters with modern-day Poles.
Didn't see that coming. Haha.
Were the proto slavic speakers then descended from a mix of Central European Celts and a Baltic like population? And are we to understand that initially some had Scythian admixture but this was subsequently mixed out of the Slavic genepool?
The Goths have very, very Scandinavian yDNA also
@Survive the Jive
Proto-Slavs definitely weren't part Celtic.
They were probably largely derived from the Trzciniec population, and then later they mixed with various Germanic groups and also came in contact with Iranic speakers.
Apart from that we need more samples from Iron Age Poland and Ukraine to see what exactly led to the formation of the proto-Slavs.
Thank you, David! Months ago together with Radek, Dr. Lukasz Lapinski and Artur Martyka we wrote something very similar with similar PCA in Polish on my blog and in the official magazine of Polish TV:
http://aldrajch.blogspot.com/2023/08/dr-ukasz-apinski-runea-ostatecznie.html
http://aldrajch.blogspot.com/2023/07/kilka-przemyslen-po-lekturze-pracy.html
https://tygodnik.tvp.pl/71930106/nasze-matki-pochodza-z-kultur-starszych-niz-rody-ojcow-jak-gotki-slowian-pokochaly
I also informed about our conclusions Prof. Figlerowicz and Prof. Stolarek. But they seem to serve Polish Autochtonism.
Chrobry: Regnum Slavorum / Gothorum sive Polonorum
Is there continuity between Trzciniec and Prezworsk? Seems fair to assume Prezworsk were Baltic groups?
This could also explain the spread of my male line R1a Y33 which is of Baltic origin and was also found in Denmark among a Viking sample on Lolland and some higher percent in Sicily, probably by the Goths or Vandals?
https://hras.yseq.net/hras.php?dna_type=y&map_type=diversity&hg=null
@ galadhorn
, thanks great article,
Regarding the Early Slavs that made it to Greece and southern parts of Balkans I believe they were for most part West Ukrainian-like but among them there probably were some South Slavic-outliers.
Unrelated but on a side note it seems Maciamo got rid all of the moderators in Eupedia and the site even started using G25. I wonder what happened?
So as I though, scy303 (Scythian outlier from Moldova) was Germanic, probably a Goth wife. She plots in the middle of the Wielbark cluster on this PCA.
@Dave
"They were probably largely derived from the Trzciniec population, and then later they mixed with various Germanic groups and also came in contact with Iranic speakers."
I agree fully, slavs could came from eastern Trzciniec, which was east of Bug.
https://i.imgur.com/0YV4Ztd.png
Polacy nic nie mają z Wielbark ?
0.021 Polak_x_scaled = 67% Poland_Middle_Ages:Poland_Ostrow_Lednicki_Middle_Ages:PCA0332 + 33% Poland_Wielbark_IA:Poland_Pruszcz_Gdanski_IA:PCA0479
0.022 Polak_x_scaled = 67% Poland_Middle_Ages:Poland_Ostrow_Lednicki_Middle_Ages:PCA0332 + 33% Poland_Wielbark_IA:Poland_Maslomecz_IA:PCA0100
0.022 Polak_x_scaled = 64% Poland_Middle_Ages:Poland_Ostrow_Lednicki_Middle_Ages:PCA0332 + 36% Poland_Wielbark_IA:Poland_Maslomecz_IA:PCA0088
0.022 Polak_x_scaled = 66% Poland_Middle_Ages:Poland_Ostrow_Lednicki_Middle_Ages:PCA0332 + 34% Poland_Wielbark_IA:Poland_Kowalewko_IA:PCA0065
0.023 Polak_x_scaled = 68% Poland_Middle_Ages:Poland_Ostrow_Lednicki_Middle_Ages:PCA0332 + 32% Poland_Wielbark_IA:Poland_Pruszcz_Gdanski_IA:PCA0498
0.023 Polak_x_scaled = 63% Poland_Middle_Ages:Poland_Ostrow_Lednicki_Middle_Ages:PCA0332 + 37% Poland_Wielbark_IA:Poland_Kowalewko_IA:PCA0063
0.023 Polak_x_scaled = 69% Poland_Middle_Ages:Poland_Ostrow_Lednicki_Middle_Ages:PCA0332 + 31% Poland_Wielbark_IA:Poland_Maslomecz_IA:PCA0114
0.023 Polak_x_scaled = 68% Poland_Middle_Ages:Poland_Ostrow_Lednicki_Middle_Ages:PCA0332 + 32% Poland_Wielbark_IA:Poland_Kowalewko_IA:PCA0040
Target: Polak_X_scaled
Distance: 1.4717% / 0.01471694
84.6 Poland_Middle_Ages
15.4 Poland_Wielbark_IA
You used only half of the 68 Wielbark samples from the Stolarek study. Therefore, on the PCA, the populations were clearly separated. But you can still see a large group of Wielbark samples located relative to the horizontal coordinate (Balto-Slavic drift) at the height of the Western Slavs and Eastern Germans.
The f3 statistics, unlike the f4 statistics, are unable to show admixture. In Stolarek's f4 statistics, most Wielbark samples share a similar quantity of origin with Northwest European and Eastern European populations.
The Balto-Slavic admixture among the Goths is best demonstrated by models using Baltic Est BA, which Arza made for the Wielbark samples published at that time. Just repeat them for new samples from the Stolarek study:
"Claims that samples from Kowalewko represent unmixed and purely Germanic population are clearly contradicted by the genomic data."
https://slavicorigins.blogspot.com/2021/05/baltic-ba-ancestry-in-wielbark-culture.html
So Davidski, are you saying Romulus is the Father of Slavs ?
Target Ukraine_Chernyakhiv.SG:MJ-19_noUDG.SG
Bulgaria_IA 9.2
Latvia_BA 16.2
Poland_Trzciniec_Culture 22.8
Poland_Weklice_WielbarkCulture_Roman.SG 18.4
Slovakia_Poprad.SG 33.4
What this shows is only at the Southeast of Poland & moving into Northwestern Ukraine, Germanic groups were mixing in with quasi-BSl populations. However, the other question is if these half breeds contributed anything to the later expanding Proto-Slavic pool.
@ambron
If the Wielbark groups were mixed then they would plot near Poland_Viking on the f3 graph.
And your point about f4 stats doesn't make any sense because f3 stats are more sensitive to recent drift.
I wonder how many years you're going to keep pretending that the Wielbark Goths are mixed before you give up?
By the way, Stolarek should retract his paper, correct it, and publish it again.
@ Romulus
''Seems fair to assume Prezworsk were Baltic groups?'
This is quite unlikely. Przeworsk is a mix of Jastorf migrants and 'Celtic' enclaves in southern regions such as Silesia. The one or two Przeworsk samples in this dataset don't support a Balto-Slavic affiliation either.
@ Rob
Thanks for that, I do vaguely remember reading about Celtic affinity in Prezworsk. There are very many outdated articles online describing Prezworsk as one of the core proto-Slavic groups.
I like how before Goths, the populations in Poland were similar to proto-Slavs.
It prevents any nihilist jerks from saying "Polish people don't belong in Poland."
They're ancestors came to Poland relatively recently but they came from Belarus which is nearby.
Usually migration and genetic change involves people from nearby areas who were similar to the people already there.
Same goes for Anglo Saxons in Britain.
They were very similar to the preexisting Bretons and didn't come from far away.
yet, nihilist anti-British people will say they were different and this makes ancient Britain "diverse."
It will be interesting to see if when we get more Gothic ancient DNA samples they will show central/east Euro admix.
It is interesting that first Goth samples are 100% Scando except for one sample.
The G2a clade in them though is not of Scando origin.
I did remove the Wielbark and Polish outliers with some Southern European and Balkan ancestry. But that was only a few samples.
@ Romulus
''Thanks for that, I do vaguely remember reading about Celtic affinity in Prezworsk. There are very many outdated articles online describing Prezworsk as one of the core proto-Slavic groups.''
It was popularized by an old archaeologist called Sedov, he's not alive anymore. But there's no basis for it, just an outdated theory.
Sometimes Przeworsk culture (PC) t is grouped into a broader Przeworsk-Zarubintsy culture homeland, as the ZC is even more commonly thought to be proto-Slavic, but that too is problematic. The ZC collapsed ~ 0 AD due to climate change and/or Sarmatian incursions, thus there is no clear link with Early Slavs (although some archaeologists juiced up some claims that there is). The PC population moved toward the Tisza region, and is associated with Vandals, and then that too disappeared by 400 AD.
Thus there is no 1:1 link between early Slavs and early Iron Age 'cultures', let alone any Bronze Age ones.
The G2a clade is Goths is under L497 which is in the same G2a clade found in Celts.
This suggests they had Celtic admixture. They don't show it in autosomes though I guess.
David
First of all, you removed 50% of the samples, including those with high Balto-Slavic genetics and Slavic haplotypes Y-chromosome, which:
"Genome-wide data suitable for further analyses (at least 10.000 SNPs for an individual for the PCA, ADMIXTURE and qpADM analyses) were obtained for 197 individuals, 68 and 129 from the IA and MA groups, respectively."
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-023-03013-9
@ambron
I ran 38 Wielbark samples in the PCA and 44 in the f3 test.
Why is it that out of all of these decent quality Wielbark samples from two papers and four different sites only one female showed Balto-Slavic ancestry?
Did you perhaps consider that the low quality samples don't really have Balto-Slavic ancestry, and instead it's just errors and contamination?
We must remember that most of those bones used to take genetic samples, were excavated, transported, analyzed by anthropologists without any precautions regarding possible contamination.
Ancient Poland--Since R1b-L51 Poland Corded Ware and R1b-L51 Afanasievo are related why not add R1b-L51 showing relative position in relation to Balti-Slavic and Weilbark cluster?
Yesterday I learned from a geneticist from Poland that the samples from Maslomecz are unreliable because they were mixed up at the stage of discovery and storage of the bones in the archive.
Maslomecz_IA looks like a Wielbark population in every sense genetically, apart from the outlier female.
But the outlier seems to be in large part of Wielbark origin too.
@Samuel Andrews
"The G2a clade is Goths is under L497 which is in the same G2a clade found in Celts."
Isn't L497 a lineage with a 7000+ BP TMRCA? I have nothing against Celtic admixture into some Germanic groups (it is probable), but this L497 is not specific enough to tell.
@Davidski
“Proto-Slavs definitely weren't part Celtic.
They were probably largely derived from the Trzciniec population, and then later they mixed with various Germanic groups and also came in contact with Iranic speakers.”
Yes, Slavs were probably largely derived from the Trzciniec. I just don’t see how you can make Slavs by mixing Trzciniec with Germanic or Baltic groups.
https://postimg.cc/BLRrqhGz
https://postimg.cc/TpsCByfk
In my opinion modern Slavs were derived from Slavic Trzciniec population which later mixed with various groups coming from the South.
Which of those Turkic Sarmatians and Scythians, with whom Slavs had contact were actually Iranic speakers?
@EastPole
Your model is purely hypothetical and unrealistic.
Only a subset of the Trzciniec population is relevant to Polish ancestry. That is, the most eastern shifted subset.
There were no migrations or any significant gene flow from the south into Poland during the Iron Age and early Medieval period.
All of the direct evidence shows cultural influences and significant population movements from the northwest into Poland at this time.
And IBD/haplotype data corroborates that Scandinavia is where the early Slavs and early Poles got their main admixture from.
@Samuel Andrews
"I like how before Goths, the populations in Poland were similar to proto-Slavs.
It prevents any nihilist jerks from saying "Polish people don't belong in Poland."
Are there really people ready to claim such things? I gather if so, it must be a nonrelevant small minority of Neonazis. In my opinion, prehistory mustn't be used to justify the expulsion of a population that has been established for few generations - and neither to claim the land possessed by another population.
@EastPole
There is no reason to assume that Sarmatians and European Scythians were Turkic speakers. They were almost certainly Iranic.
(I am less sure about the language of "Scythians" around the Altai, but that is a different story, because Iron Age proto-Slavs had no contact with those.)
@Samuel Andrews
On a related note, recently I read about a new archaeological paper showing that the Helvetii arrived in what is now Switzerland much later than previously thought: in the first century BC, just a few decades before they attempted to migrate on to what is now France. Leftist-liberal newspapers concluded that thus, the Helvetii were not really our primordial forebears. Alternatively, they might have highlighted that we are descendants of migrants, but they didn't. In any case I commented to their article that, since the Romans forced them to return to what is now Switzerland, they may be regarded as our forebears anyway, with limitations.
@ EastPole and Slumbery
Scythians were Indo-European-speaking Iranian nomads who were later largely absorbed by Turkic-speaking nomads of Eastern Europe and Central/East Asia. In Europe, some traces can be found, for example, among today's Romanians and in a diluted form among Poles from the Carpathian Basin through return migration of Ostrogoths (contact with Alans)
@Davidski
“Your model is purely hypothetical and unrealistic.”
From Wikipedia:
“The Lusatian culture developed as the preceding Trzciniec culture experienced influences from the Tumulus culture of the Middle Bronze Age, essentially incorporating the local communities into the socio-political network of Iron Age Europe.”
“The Tumulus culture was eminently a warrior society, which expanded with new chiefdoms eastward into the Carpathian Basin (up to the river Tisza), and northward into Polish and Central European Únětice territories.”
Migrations from the Carpathian Basin to Poland made the Trzciniec population more similar to modern Slavs.
This model is quite realistic:
https://postimg.cc/G9qPXDSs
Your model is purely hypothetical and unrealistic. How do you imagine it demographically? You know Goths and Balts were very small populations and Slavs were huge populations, as they lived on the best agricultural land in Europe. So how did it happen? Do you think you can take 10000 Goths and 10000 Balts and make 10,000,000 Slavs in a few generations? And how did it happen? Were Balts actually stealing women from Goth’s villages?
@Davidski:
“And IBD/haplotype data corroborates that Scandinavia is where the early Slavs and early Poles got their main admixture from.”
Are we able to cut to the chase here? Are you saying that Poles are a mix of two groups:
(1) “true” Slavs from Belarus (you sort of agree on Trzciniec - except you keep emphasizing ‘eastern’ Trzciniec which sounds like you want to deephasize western Trzciniec) except that you can’t quite say this yet bc we don’t have BA/IA samples from the Belarus/Ukraine border or from Belarus?
And note we also don’t have such samples from Western Trzciniec or Przeworsk…
And (2) some Scandinavian population?
@Davidski
I would like to ask about 3 things:
a. since you have now analyzed the topic thoroughly, please tell me whether your opinion about the PCA0475 sample (Z2103-CTS7822) has changed in any way? Do you still think this is a Scandinavian sample?
b. what do you think about the PCA0300 sample dating back to the 10th century? It also has Z2103 and then By593.
c. Do you have any opinion about iwno culture? Do you see more references to Trzciniec or Unetice culture in it?
In reference to other people's statements, it hurts my head when I read "Slavic Lusatian culture", "Slavic Przeworsk culture", etc. I understand being fascinated, but such fanaticism is at least strange.
In my opinion, the name "Lusatian culture" is quite incorrect. All its groups, apart from some common features, were fundamentally and very different. The Western Lusatian culture and the Eastern Lusatian culture could as well be different cultures. The western one is based on a completely different background, more related to the areas of Germany, etc., so we can say that Slavs were present in the eastern part of the Lusatian culture... but let's not make jokes by writing that the entire area was Slavic :)
I think the G2a is a generic central European clade, although it might fall under 'Celtic'. Probably more to do with Urnfield influences on the Pomeranian culture.
@Dranoel
PCA0475 still looks Danish. That's unlikely to change.
I'm not familiar with PCA0300. I only have the relatively high coverage samples.
I don't have any opinions about the Iwno culture. I need to see some samples from it.
@Lucas
Proto-Slavs formed in large part from a subset of the Trzciniec population that was Baltic-like, and thus eastern genetically.
Whether this means that this Trzciniec or Trzciniec-derived group was also eastern geographically is still an open question. Lots of options here, don't you think?
https://i.imgur.com/0YV4Ztd.png
But it's likely that there were other peoples involved in the formation of the early Slavs, probably including the Wielbark Goths. Hopefully more samples from the Iron Age and early Middle Ages will help to clarify this.
Poles are largely derived from proto-Slavs, but with some extra Germanic admixture from the Wielbark Goths, Vikings, recent German migrants and so on. There might also be some significant West Baltic ancestry in parts of Poland today.
Indeed, gradual gene flow into the ethnic Polish population from basically all of the populations living in and near Poland seems to have had a minor but noticeable impact on the Polish gene pool and the shape of the Polish cluster in my PCA plots.
@Davidski:
Thank you. BTW, where does that Trzciniec circle picture come from? There are other pictures of Trzciniec’ extent that have it as more western and southern. I suppose it may depend on the period in question.
No idea where it was published. I got it from here.
https://eurogenes.blogspot.com/2023/11/wielbark-goths-were-overwhelmingly-of.html?showComment=1699637602612#c5588035958175168364
Seems more or less accurate.
That image is from Barrows, Long-distance Mobility and Peripheral Elites between the Baltic and Black Seas during the 2nd Millennium BC
But obviously the adna doesn’t support the view that these groups were migrants from the steppe, as I said before
Thanks. Yes, it’s from Makarowicz.
Dave - i think the main Wielbark cluster can be modelled as a 2 way mix of La Tene (e.g. Czech) & Swe-IA
That's probably because the Goths were more Danish-like than Swedish.
@Dranoel
Polski egzemplarz R1b-Y14300
0.023 R1b_14300_scaled = 55% HRV_Bezdanjaca_BA + 45% Baltic_Bronze_Age:_LVA_BA
0.024 R1b_14300_scaled = 56% HRV_Bezdanjaca_BA + 44% Baltic_Bonze_Age:EST_BA
(-)
0.035 R1b_14300_scaled = 298% Unetice:CZE_Unetice_C - 198% Bell_Beaker_England
0.037 R1b_14300_scaled = 172% Tumulus_Culture:CZE_MBA_Tumulus - 72% Spanish_Late_Bronze_Age:ESP_LBA
Target: R1b_14300_scaled
Distance: 1.5673% / 0.01567346
26.4 Baltic_Bonze_Age
20.8 Baltic
19.8 Eastern_Corded_Ware_Srubnaya
13.8 Vucedol
12.4 BGR_ChL_outlier
2.4 JOR_PPNC
1.8 SRB_Yamnaya_EBA
1.4 MDA_Catacomb_Danceni_MBA
0.8 North_Cushitic
0.4 Japan_&_Korea
Here is Makarowicz paper about Trzciniec https://repozytorium.amu.edu.pl/bitstreams/cd67a0bf-298a-49d7-b2ec-de26a2797f40/download
G25 based PCA made by Ph2ter form Genarchivist forum. Clearly we see Trzciniec samples were in different cluster then MA Slavs.
https://i.imgur.com/Vc09vyK.jpeg
David, there is too much evidence of the genetically mixed Scandinavian-Slavic nature of the Wielbark Goth population to dismiss it all as errors or contamination.
But I don't want to argue with you about this, because ultimately my conclusions regarding the biological history of the Polish population are similar to yours.
I would like to point out that in ethnolinguistic research, genetic data cannot be analyzed in isolation from linguistic facts. And linguistic facts (the oldest layer of Slavic toponymy) show that in the Trzciniec area we can only find the Proto-Slavic homeland in the zone between the middle Warta and the middle Dniester.
We need only pure logic to assume that eastern genetically part of Trzciniec people must be derived from eastern paret of this culture, not western, othwerwise genetically western-like would be live in Belarus or Ukraine? Absurd:)
@Lukaszer
What are the earliest examples of Slavic speakers that we have in the G25?
Can you make a list with their C14 dates and locations?
@ Davidski
''That's probably because the Goths were more Danish-like than Swedish.''
Dont think so, I used Denmark_IA as well as SWE_IA.
But it was only a quick look.
I don't think there is any evidence for the main wielbark cluster having recent slavic admixture, unless you also think Denmark_IA/Sweden_IA have it.
@ Polak_X
Thank you very much!
@ Davidski
Given the data provided by Polak_X, are you able to say anything more about this sample? What is its character and what population does it seem to be similar to?
Łukaszer
So you have to come to terms with the fact that the genetically eastern part of Trzciniec inhabitants coming from Poland is an absurdity that defies logic.
C14 oldest Slavs:
HUN_Avar_Szolad__Av1_590
HUN_Avar_Szolad__Av2_590
CZE_Early_Slav__RISE569_715
POL_MA_Piast__PCA0143_874_R1a_M458
CZE_MA_(not)Knoviz__I20515_942
POL_MA_Piast__PCA0156_949_I2_Y3120
May be Slavs C14:
TUR_Marmara_Ilipinar_Byz3__I10430_732_I2_Y3120
HRV_Trogir_Byz_o__I15742_718
Arch. context oldest Slavs:
CZE_MA_Pohansko__POH11_850_R1a-CTS1211
CZE_MA_Pohansko__POH13_850
CZE_MA_Pohansko__POH27_850
CZE_MA_Pohansko__POH28_850_R1a
CZE_MA_Pohansko__POH3_850
CZE_MA_Pohansko__POH36_850_I2_Y3120
CZE_MA_Pohansko__POH40_850
CZE_MA_Pohansko__POH41_850_I2_Y3120
CZE_MA_Pohansko__POH44_850_I2_Y3120
POL_MA_Piast__PCA0115_950
POL_MA_Piast__PCA0119_950
POL_MA_Piast__PCA0122_950
POL_MA_Piast__PCA0124_950
POL_MA_Piast__PCA0126_950
POL_MA_Piast__PCA0127_950_I2_Y3120
POL_MA_Piast__PCA0130_950
POL_MA_Piast__PCA0132_950_I2_Y3120
POL_MA_Piast__PCA0133_950
POL_MA_Piast__PCA0137_950
POL_MA_Piast__PCA0138_950
POL_MA_Piast__PCA0140_950
POL_MA_Piast__PCA0142_950
POL_MA_Piast__PCA0146_950
POL_MA_Piast__PCA0148_950
PCA (in red squares C14 dated and others in pink colour):
https://i.imgur.com/Ok3V8wx.png
@Dranoel
I don't have a clue what Polak_X is talking about.
EthanR
Swedish IA has a recent Slavic admixture from Poland, as Margaryan presents in his work. It is younger than the Wielbark admixture. All this can be clearly seen in old analyzes of Arza, where the native Scandinavians have no Balto-Slavic admixture:
Baltic EST BA:
Kow 54 – 26.8
Kow 29 – 24.4
Kow 26 – 19.6
SWE Oland IA VK579 – 18.4
Kow 22 – 16.8
Kow 25 – 16.6
SWE Oland IA VK522 – 15.2
DNK Sealand IA VK532 – 11.8
DNK Sealand IA VK213 – 0.0
DNK Jutland VK582 – 0.0
https://slavicorigins.blogspot.com/2021/05/baltic-ba-ancestry-in-wielbark-culture.html
There's no Slavic admixture in Sweden_IA and Margaryan never claimed such a thing.
Dranoel-Goths were more Danish-like than Swedish
Polish/Slavic Y14300 Yamnaya like-derives from R1b-L23+, found in samples like Yamnaya-I0443-found so far found the mostly homogeneous paternal cluster associated with 300+/- wagon burials, early development of Dom2, and iron burial samples. It remains to be seen if the Russian samples of R1b from Abashevo -Pepkino burial mound(1 blacksmith) were Z2110 or Z2109 and if Z2110 is found in Hungarian Yamanaya and or Catacomb culture.
https://eurogenes.blogspot.com/2020/06/the-abashevo-axe-did-it-mednikova-et-al.html
However the Yamnaya like samples with R1b-Z2110+ have not been found in the homogeneous Yamnaya-Afanasievo cluster comprised of R1b Z2109+and L51+ 310+, Or Hungarian Bell Beaker I0744 and Beaker_Hungary_I2787.
Beaker_Hungary_I2787
Beaker_Bavaria 0.442±0.045
Yamnaya_Samara 0.558±0.045
chisq 8.562
tail prob 0.73982
Full output
Beaker_Hungary_I2787
Beaker_Czech 0.441±0.045
Yamnaya_Samara 0.559±0.045
chisq 10.009
tail prob 0.61513
Full output
Beaker_Hungary_I2787
Beaker_The_Netherlands 0.576±0.062
Yamnaya_Samara 0.424±0.062
chisq 11.469
tail prob 0.489238
https://eurogenes.blogspot.com/2019/01/hungarian-yamnaya-bell-beakers.html
https://bellbeakerblogger.blogspot.com/2017/07/szigetszentmiklos-cemetery-santas-six.html
PCA0475 sample (Z2103-CTS7822)
PCA0300 R-V2986
Polish
https://www.yfull.com/tree/R-Y14300/
Maros samples
Earliest Z2110 CTS-7556
https://www.yfull.com/tree/R-CTS7556/
Viking
https://www.yfull.com/tree/R-Z2108/
VK535, Viking found in Italy.
David
Indeed, "Slavic admixture" is my simplification. Sweden IA has a recent Baltic BA admixture, and Margaryan's fineStructure shows that it is about ancestors coming from Poland.
Sweden IA doesn't have any Baltic admixture, nor any ancestors from Poland.
Baltic and Slavic admixture starts showing up in Scandinavians during the Viking Age.
David
There is no point in arguing with the facts:
"The allele frequency in Iron Age samples is at intermediate levels (c. 37.5%), suggesting this rise in frequency must indeed have occurred during the Iron Age (c. 1500-2500 years ago), but was largely complete at the onset of the VA. Interestingly, the allele frequency of the allele is much higher (c. 40%) in the Bronze Age population from the neighboring Baltic Sea region than in Bronze Age Scandinavia. Given the geographic and cultural proximities between Scandinavia and the Baltic region, this may suggest gene flow between the two regions resulting in increased frequency of lactase persistence in Scandinavia during the Iron Age."
https://www.biorxiv.org/content/10.1101/703405v1.full
@ambron
They're talking about the lactase persistence allele, and speculating that the allele rose in frequency in Scandinavia because of admixture from the East Baltic.
This is funny, because there's no direct evidence of admixture from the East Baltic in Bronze Age or Iron Age Scandinavia. Not even in this paper.
Also, it's been shown that this allele rose in frequency due to selection not admixture.
OMG I've never imagined Ambron is talking only about lactase persistence allele, not whole autosomal admixture nn relation to his hypothetical "BaltoSlavs in IA Scandinavia":) I have no words.
.........................
.........................
Target: SWE_BA:RISE175
Distance: 1.6547% / 0.01654696
28.2 RUS_Sintashta_MLBA
22.0 Baltic_EST_BA
13.8 Bell_Beaker_England
11.8 Iberia_Southeast_BA
8.4 England_CA_EBA
5.0 RUS_Srubnaya_MLBA
2.8 FRA_Occitanie_EMBA
2.6 DNK_LN
2.4 MAR_EN
1.8 MNE_Vrbička_N
0.8 Rochedane
0.4 DEU_Wartberg_MN
Interestingly, Makarowicz is also listed on the Chyleński paper. Presumably to make this interdisciplinary.
@Lukaszer:
I find that PCA a bit confusing. It has a cluster for IA Slavic ancestors with samples including Sicily, Gotland and Russia? Is that even a cluste
Lukaszer
You're embarrassing yourself once again.
Do you know of any allele that passes from parent to offspring without all the inherited genetic material flowing?
David
Baltic EST BA:
Kow 54 – 26.8
Kow 29 – 24.4
Kow 26 – 19.6
SWE Oland IA VK579 – 18.4
Kow 22 – 16.8
Kow 25 – 16.6
SWE Oland IA VK522 – 15.2
DNK Sealand IA VK532 – 11.8
DNK Sealand IA VK213 – 0.0
DNK Jutland IA VK582 – 0.0
In each of these cases, can the Balto-Slavic admixture be explained by error or contamination?
Ftdna has uploaded the Trzciniec samples, looks like they were able to get further downstream calls on many samples for Y DNA. I have 9 new matches from them showing L233+.
@ambron
These samples don't have any Balto-Slavic admixture.
Baltic EST BA is just being used as a proxy for local Scandinavian ancestry.
Ambron should be careful and come to terms with the results. Otherwise he'll end up like that psuedo-linguist Jaska and his 3 loser fanboys on GeneArchiver
Balto-Slavic+Proto Indo-Iranian
Afanasievo + Polish + Czech Corded Ware L51+ and Czech Corded Ware R1b-Z2103+ might have been able to communicate with Abashevo-Sintashta R1b-Z2103+ about Dom2 horses from Turganic region used in Chariots, no? .
Corded Ware cultural and genetic complexity (Linderholm et al. 2020)
https://eurogenes.blogspot.com/2020/04/corded-ware-cultural-and-genetic.html
Tracing its origins in the Fatyanovo–Balanovo culture, an eastern offshoot of the Corded Ware culture of Central Europe, the Abashevo culture is notable for its metallurgical activity and evidence for the use of chariots in its end phase + Indo-Iranian-speaking or Proto-Indo-Iranian-speaking
@Romulus
How do you access these ftdna samples?
@Lucas
https://discover.familytreedna.com/y-dna/I-L233/tree
@Ethan:
Thanks. Looks like they now have two 1211s under 280 (raciborowice kolonia 794 and pielgrzymowice 711). Under m458 they have a few kowalewkos (2, 38, 54) and czarnówko 547.
@Lucas
What are the C14 dates and contamination levels of these M458 Wielbark samples?
Lucas
Kow 54 - M458, 26.8 Baltic EST BA
But your effort is in vain, because David will tell you anyway that it's either an error, contamination, or a Scandinavian proxy.
Lucas
Kow 54 - M458, 26.8 Baltic EST BA
But your effort is in vain, because David will tell you anyway that it's either an error, contamination, or a Scandinavian proxy.
There's no M458 or Balto-Slavic ancestry in any of the decent coverage Wielbark samples with low level contamination.
If you're forced to prove something so obvious as the mixed nature of the Wielbark population with barely usable samples, then you have to at least acknowledge that you have a problem.
@Lucas Slawbuski to @Lukaszer:
"I find that PCA a bit confusing. It has a cluster for IA Slavic ancestors with samples including Sicily, Gotland and Russia? Is that even a cluster"
This is my PCA. The samples from Sicily are military mercenaries with Balto-Slavic autosomal and with Y-DNA common to Trzciniec samples (I2-L233 and R1a-CTS1211).
Sample from Russia is actually a sample from Iron Age Ingria, also with Balto-Slavic autosomal makeup.
Gotland is Medieval Viking sample with R1a-M458 Y-DNA. It is in this cluster because it has Baltic autosomal.
These samples form a cluster because they are the only Iron Age samples inside Polish Medieval cluster. They all are similar to the Slavs autosomally and some also by their YDNA. They can be treated as an eastern continuation of Trzciniec cluster.
Here in this PCA they are marked with red square:
[img]https://i.imgur.com/o6dgTr0.png[/img]
Do you have any ideas about it?
https://imgur.com/Gf5o7cL
Daviski:
"Baltic EST BA is just being used as a proxy for local Scandinavian ancestry."
How this understand? Should I understand this to mean that Poles (47.0 Baltic EST BA) derive most of their ancestors from Scandinavia?
I can, of course, agree that Baltic BA at the level of several percent may be hum resulting from the distant common origin of Scandinavians and Balto-Slavics, but not at the level of a dozen or several dozen percent.
Of course, I admit that I have a problem, because the problem is archaeogenomics itself. However, just because a sample does not meet the SNP limit for G25 does not automatically mean it is an error or contaminant.
Target: Wielbark_Kowalewko:PCA0027
Distance: 2.4836% / 0.02483641
56.6 German_Frisian
32.0 Latvian
7.6 Basque_French
1.8 Igorot
1.2 Finnish_Southeast
0.8 Ju_hoan_North
@ambron
Data quality is a very important issue that doesn't get enough attention in ancient DNA papers, let alone among online hobbyists.
I generally remove samples from the G25 when they have less than 10% of the SNPs available.
But even at 10-15% many samples produce noisy, uncertain results.
This isn't usually a huge problem when studying basic, broad ancestry components, but it can be a serious issue when looking at fine-scale ancestry, like intra-European genetic components.
And there's no way that anyone can reliably work out with f4 stats whether a low coverage sample like that is North Germanic or Balto-Slavic, or a mix of the two.
There should be a higher threshold in ancient DNA papers for missing data and contamination, especially in certain studies that focus on fine-scale ancestry.
You and many others here need to start thinking about this issue instead of pretending that there's no difference whether a sample has 15,000 markers or 1,000,000 markers.
You need to start paying attention both to the number of markers available and the contamination levels.
@dancingfragments
Your models are statistically overfitted and they don't reflect reality.
Davidski, see graph
https://imgur.com/a/qVnSKa6
Target: Wielbark_Kowalewko:PCA0027
Distance: 4.4446% / 0.04444641
31.2 Baltic_EST_BA
29.2 Yamnaya_RUS_Samara
25.6 TUR_Barcin_N
12.8 WHG
1.2 CHN_Yellow_River_LN
@dancingfragments
There were definitely contacts between Germanic groups and steppe nomads, so this might be what you're seeing there, and the Asian ancestry might be real.
I've done a few blog posts about this.
https://eurogenes.blogspot.com/2019/07/asiatic-east-germanics.html
https://eurogenes.blogspot.com/2018/03/exotic-female-migrants-in-early.html
Davidski, 1st Century BC by Tomenable
Target: Wielbark_Kowalewko:PCA0027
Distance: 2.7783% / 0.02778302
74.4 Europe_Celto_Germanic
23.2 Europe_Balto_Slavic
2.2 Asia_Ainu_People_Japan
0.2 Africa(South)_Khoisan_Peoples
Target: HUN_Avar_Szolad:Av2 ( To jest to co przypomina znalezisko zespółu Poznańskiego )
Distance: 1.5312% / 0.01531176
40.2 Belarusian
32.8 Polish
25.0 Russian
2.0 Irish
Target: HUN_Avar_Szolad:Av1 ( Każdy Wielkopolan Kujawiak ma tę osobę na pierwszym miejscu)
Distance: 0.8262% / 0.00826234
33.6 Polish
32.6 Ukrainian
20.4 SarmatianScythian
8.2 Russian
5.2 Irish
This sample might have some eastern ancestry, so the Asian admixture might be real.
But if it is, then the eastern ancestry is from somewhere like Ukraine, Russia, or the Balkans, not from supposedly Iron Age Polish Slavs.
See here...
https://eurogenes.blogspot.com/2019/07/asiatic-east-germanics.html
arget: Poland_Kowalewko_IA:PCA0027
Distance: 2.0002% / 0.02000225
34.8 Poland_Kowalewko_IA
26.4 Poland_Maslomecz_IA
16.4 Poland_Niemcza_Middle_Age
14.8 Poland_Ostrow_Lednicki_Middle_Age
4.2 Poland_Lad_Middle_Age
2.4 Poland_Santok_Middle_Age
1.0 Poland_Pruszcz_Gdanski_IA
Target: Poland_Kowalewko_IA:PCA0027
Distance: 1.8822% / 0.01882213
52.2 Poland_Kowalewko_IA
23.2 Poland_Trzciniec_Culture
7.8 Poland_Ostrow_Lednicki_Middle_Age
7.6 Poland_Niemcza_Middle_Age
4.4 Poland_Santok_Middle_Age
3.6 Poland_Milicz_Middle_Age
1.2 Poland_Maslomecz_IA
Target: Sweden_IA.SG:RISE174_noUDG.SG
Distance: 1.2934% / 0.01293429
63.2 Poland_Kowalewko_IA
26.6 Poland_Trzciniec_Culture
6.2 Poland_Niemcza_Middle_Age
1.8 Poland_Maslomecz_IA_oEastEurope
1.4 Poland_Maslomecz_IA
0.8 Poland_Legowo_Middle_Age
Target: Sweden_IA.SG:RISE174_noUDG.SG
Distance: 1.4032% / 0.01403230
61.2 Poland_Kowalewko_IA
11.8 Poland_Niemcza_Middle_Age
10.0 Poland_Legowo_Middle_Age
10.0 Poland_Maslomecz_IA
2.8 Poland_Gaski_IA
2.6 Poland_Rumin_Middle_Age
1.6 Poland_Maslomecz_IA_oEastEurope
Ambron, I think Goths were mixed with Slavs, even some Goths in Sweden. There was trade over the Baltic, so it is natural.
@ Dawidski
Myślę że ważną miejsce dla Słowian i Polaków odegrała Nizina Węgierska .
Distance to: Polak_X_scaled
0.02522712 HUN_Avar_Szolad:Av1
0.03223790 Magyar_West_Eurasian:HUN_Conqueror_elite_AGY75
………………
0.04017612 HUN_Avar_Szolad:Av2
Podobny dystans ma @ ph2ter Chorwat.
Goths and other east Germanics were mixed with Huns and other really eastern peoples from far Eastern Europe, and some of these mixed Goths moved back west.
Not only has this been proven with ancient DNA already, but it's historically documented, and we discussed it on this blog years ago.
Myślę że ważną miejsce dla Słowian i Polaków odegrała Nizina Węgierska.
Bullshit.
I greet you, dear Davidski, I really respect you for your work and your own calculator, I have a question, if there is some special calculator where you can make a mixture of components yourself (I want to add ANE to CHG in different proportions and see what result will be closer to which nations ) or subtract some components from others
forgive me for my English, I'm using a translator
Just combine the coordinates from the relevant ancient samples in the desired proportions into one population or one population average.
To do that you would have to rename the coordinates accordingly and learn how to create population averages.
I don't have time to write up a guide but many people have been doing this for a while.
@Polak_X
Myślę że ważną miejsce dla Słowian i Polaków odegrała Nizina Węgierska
Hungary?
I thought M458 Balto-Slavic and Fatyanovo origin were connected to the oldest Corded Ware sites. Sites like Denmark(SGC) or Lesser Poland or Sredny Stog/Deriivka ? Has this changed?
@Davidski
Unrelated, but there seem to be some issues with a minority of samples in the new G25 sheets. For example these 2 Amur river_EN averages:
China_AmurRiver_BA,0.0387,-0.239665,-0.067505,0.032623,-0.023389,0.041834,0.005875,0.006692,0.020248,0.024602,-0.041896,0.003447,0.006838,0.005918,-0.008415,-0.007955,-0.008605,-0.001774,-0.000503,-0.000625,-0.008235,0.016569,0.008381,-0.003253,0.01892
China_AmurRiver_EarlyN,0.035285,-0.239665,-0.081458,0.04522,-0.023235,0.039463,-0.00235,0.0099225,0.0307805,0.023782,-0.040597,-0.004796,0.010481,-0.0024085,-0.009704,-0.0076235,-0.0007825,-0.0088045,-0.012444,0.003439,-0.0008735,0.0175585,0.00228,0.003735,0.007424
When run in G25 with the appropriate references that work on the rest of Siberians/Northern East Asians these two get very bad fits and about half of their ancestry comes up as SIS_BA2/South-Central Asia when I by chance left that in the model. The older coords of the same individuals do not show this and I also noticed that you have them labeled as genotyping_noUDG could the first part stand for genotyping error of some sorts?
okay, let's leave this topic, I'll sort it out myself, I noticed that there are often posts here on the topic of the homeland of the Proto-Indo-Europeans, or those CHG that were given mixed with EHG to the Yamnaya people , and I would like to insert my two cents at this point, for some reason It seems to me that the true place of contact between CHG and EHG was the Eastern Caucasus - the Caspian lowland, I myself come from Dagestan, our languages do not belong to the Indo-European group, but at the same time the peoples of Dagestan have one of the highest yamnaya components in the world and the distances are low (especially the Dargins Akhvakhians Kubachi residents Kaitags Avars and so on), we have so many languages due to the fact that people have been living for quite a long time and isolated from each other, it is archaeologically and genetically confirmed that the ancestors of the Dagestanis live in the mountains at least 10 thousand years (Chokh Mesolithic), so here I am saw a theory on the Internet that there was an exchange of women between hunter-gatherers from Dagestan and Eastern European hunter-gatherers who came from the north, do you think this theory has a right to life? you’re probably tired of everyone trying to cling to the history of the Proto-Indo-Europeans)
@Davidski:
Don’t know. All I was pointing out was that they made it to ftdna. Obviously if they were contaminated or misdated from the start that’d be a problem. That said, not sure how readers can judge contamination or bad dating from the paper itself? Or are you saying you ran these through your traps and you are showing contamination/bad c14? (TBC, I am not yet far enough down the rabbit hole to be running own PCAs, etc.).
@ph2ter:
Thank you.
“These samples form a cluster because they are the only Iron Age samples inside Polish Medieval cluster. They all are similar to the Slavs autosomally and some also by their YDNA. They can be treated as an eastern continuation of Trzciniec cluster.”
Still, the above is strange.
EastPole
Despite David's skepticism, it is obvious to me that the Wielbark Goths have a Slavic/Balto-Slavic admixture. The IA Swedes also have a Slavic/Balto-Slavic admixture, as I was assured by the critics of Stolarek's work (including one academic geneticist), who cited this fact to prove that the Goths could have brought this admixture with them from Scandinavia.
David
Your comments about the quality of the samples are of course correct. However, in the scale of the entire population, lower quality samples are equally important because they show the frequencies of specific characteristic alleles occurring in it.
@ all
and not to forget the two Celtic guys with East Asian input, Bylany_D112_Hallstatt
Is there any way to figure out and isolate modern percentages autosomally of Gothic ancestry in modern day Europeans. I1 is all over Europe and it’s a clear indicator of Gothic migrations in eastern and southern Europe.
@Lucas
The coverage and contamination levels for these samples are listed in a table in the paper's supplementary file.
The Wielbark M458 samples are indeed of low coverage and they do show relatively high contamination.
So it's possible that the M458 markers actually belong to archeologists or some other people who handled the remains.
@Арсен
To support David's direction to average your samples, if you want a headstart this site has a simple tool to get you started.
https://www.exploreyourdna.com/average-g25.aspx
For more advanced mixes, you'll want to learn to manually calculate them.
HTH
@Davidski
Understood. Presumably contamination should go hand in hand with misdating, i.e., assume they have not contaminated these with an IA 458 from some other location. In any event, would think that would be highly embarrassing for our authors?
@Lucas
Considering the lax peer review in ancient DNA papers from peer reviewers who don't know much about fine-scale ancient DNA analysis, it's pretty easy to create an impression of two distinct Northern European populations being closely related and even mixed.
If you're from a half decent scientific institution, you can get your paper published and it'll take years before anyone challenges the findings, if ever.
Meantime, other people are free to cite and quote the work as a peer reviewed source to back up their own funny ideas.
No one's embarrassed about anything, as far as I can see.
@gamerz_J
Garbage samples. I'll remove them.
@ Davidski
None of these Goths have eastern ancestry. Why are you taking those garbled, anachronist models seriously ?
They are mostly a two-way mix of Nordic and Central Europe, eg.
Poland_Kowalewko_IA:PCA0030
Czech_LBA_Knoviz 45.0
Denmark_LN.SG 41.8
Latvia_BA 12.4
d 0.02
The LAtvia_BA might be noise eventually Ill check with qpADM
Goths only acquired eastern admixture when they reached the Black Sea steppe
@ Steppe
there's no real eastern Scythian ancestry in Czech Hallstatt
Czech_IA_Hallstatt.SG
Czech_LBA_Knoviz 97.6
Most of these Wielbark samples are really of very poor quality, and with no UDG treatment.
So there's all sorts of noise there, and it's best to combine them into population sets.
When we do that they generally come out more western than Scandinavian Vikings.
https://blogger.googleusercontent.com/img/b/R29vZ2xl/AVvXsEjVOAGHspJcbOFh5gbhwYjKbqie5ys6nHjeNhWRSXESjTD1kMNf5sB-NsMXnNORnu9VC4190FyV7Stt38jM7WJbH1MzxArtxpcVhgU62bgozGeGiGp31JzI3_laVzQ0eaYkwdxN-9mq2BKdE14AmeM60WoKBCZ1Js_9Zc7QAyHqc113Q_sJd3zX_ZpryeYN/s1279/Poland_Wielbark_IA_f3_graph.png
hhhm yeah, on PCA they show extreme drift, close to Vikings. Doesn't accord with the G25-based admixture inferencing.
It's strange because the old versions of those samples are okay
CHN_Amur_River_9900BP:NE30,0.020488,-0.450895,0.070144,-0.054264,-0.038161,-0.045459,0.006345,0.013846,-0.001841,0.023508,-0.044657,0.004046,0.001933,0.003165,-0.007193,0.006364,-0.00352,0.010515,0.010056,0.023011,0.010482,-0.019413,-0.016022,-0.001687,-0.022633
CHN_Amur_River_8900BP:NE35,0.012521,-0.450895,0.072784,-0.050388,-0.047086,-0.044065,0.012926,0.015461,0.009204,0.023873,-0.046443,3e-04,-0.003122,0.001514,-0.005022,-0.009679,0.006258,0.00152,0.004274,0.011631,0.018343,-0.024978,-0.018364,-0.001928,-0.014849
@Rob
Hallstatt_DA112:
mtDNA: HV0+195 has an East Eurasian input just like the one sample from IA_Prescythian_Mezocsat, there were contacts between the Hallstatt zone and the steppes (Cimmerians and Scythians) this is also shown by archeology as in the book “Riding Nomads in Europe”
No
„Myślę, że miejsce wystąpienia dla Słowian i Polaków zagrała Nizina Węgierska”
Should
Myślę_że_ważnym_miejsce_dla_Słowian_i_Polaków_była_Nizina_Węgierska.
Przepraszam
Lucas
In fact, the most suspicious are samples that are too good quality. Because the degradation of biological material progresses over time, a sample of too good quality raises the suspicion that its DNA is a contamination from a person buried later or from a scientist examining the bone material (archaeologist, anthropologist or geneticist). Such a sample is usually excluded from further analysis at the very beginning. It can also be subjected to verification procedures, such as C14 dating, molecular dating with subsequent mutations in the Y chromosome and confrontation with mutations in the Y chromosome from scientists who had contact with bones and biological material.
Because the Y chromosome makes up only 0.38% of the genome, its characteristic polymorphisms may be preserved in their entirety (or almost in their entirety) even in low-quality samples. Wielbark M458 samples clearly show signs of degradation of biological material over time and do not contain young polymorphisms - medieval or modern. In my opinion, they are further evidence of the mixed Scandinavian-Slavic genetic character of the Wielbark Goths.
@ambron
In fact, the most suspicious are samples that are too good quality.
Such a sample is usually excluded from further analysis at the very beginning.
This is pure fiction. There has never been a single case of this happening.
High coverage ancient samples with low levels of contamination are never excluded from studies.
Obviously, the samples that are too heavily damaged and/or contaminated are the ones that are routinely removed from further analyses.
From this point, I would urge anyone reading these comments to ignore anything that ambron has to say about ancient DNA.
@ steppe
That reference is rather generic. The “Scythians” in Central Europe weren’t easterners
On my admittedly quick look, Halstatt does not seem to have of the sort of ancestry seen in IR1. Little wonder , he’s R1b-U152
But im interested to see how you might demonstrate shamanka or Altai related ancestry there ?
I have simplified this problem to make it easier to understand for those less familiar with genetics. To clarify:
Estimating the level of contemporary DNA contamination during screening is crucial when deciding whether to include a sample in further analyses.
@ Rob
ask David or Ph2ter they make the calculator and then you see the input
@ Steppe
''ask David or Ph2ter they make the calculator and then you see the input''
The Bylany individuals plot within the Knoviz & Czech Tumulus Culture cluster, if anything theyre slighty west shifted
Check it for yourself
People might have thought there is some link with the east, but its rather western Scythians which have Halstatt ancestry.
@Rob
D112 Hallstatt Bylany has a slight East Asian input autosomal and the mtdna is specific!
DA112 has negligible East Asian admixture but its mtDNA is perfectly European. HV0-T195C! is mostly western European clade present in Europe at least since Neolithic.
DA112
-------------- ANCESTRY BREAKDOWN: -------------
18.743% HUN_LBA__I20751_-1250
17.255% CZE_LBA_Knoviz__I15959_-1050
13.644% HUN_LBA_Halva__I20749_-1250
13.284% CZE_LBA_Knoviz__I13793_-1050
9.021% DEU_Tollense_BA__WEZ15_-1200
7.819% DEU_Tollense_BA__WEZ58_-1200
6.358% NLD_MBA__I26831_-1450
4.553% DEU_Tollense_BA__WEZ53_-1200
2.794% DEU_Tollense_BA__WEZ61_-1200
2.350% CZE_LBA_Knoviz__I20526_-900
2.107% DEU_Singen_EIA__MX265_-597
1.639% MNG_Ulaanzuukh_Slab_Grave__BOR001_-903
0.432% FRA_Alsace_IA1__Jeb8_-500
------------------------------------------------
Fit error: 0.02453171773441679
@Davidski,
you also made an article about the two Hallstatt samples D111 and D112 back then?
@ ph2ter
Thank you, it's all good and IA IA_Prescythian_Mezocsat?
Ambron does have a point though. If there are pristine samples that suggests contamination more than crappy samples do - though crappy samples may suggest, well, crappiness. It would seem, if there are questions, a relatively simple exercise to redo these. Of course that won’t stop intentional “mistakes” if that’s where we are heading here.
It's funny the Wikipedia article titled "Origin of the Goths" starts out "Concerning the origin of the Goths before the 3rd century, there is no consensus among scholars." and cites two sources published fairly recently. The Goths are exactly who the Roman writers said they were.
@Rob
DA111 plots within Modern French and Ancient Gaul in Dave's own Europe PCA.
@Lucas Slawbuski
ambron doesn't have a point.
In his panic to try and save his pet theory he confused two issues:
- ancient DNA damage patterns that help to identify ancient sequences and differentiate them from modern DNA
- the high quality of a high coverage ancient sample.
High coverage ancient samples are often of higher quality than modern samples, but no one in their right mind would ever confuse this high quality with contamination, because there are very specific ways to work out modern contamination in ancient DNA.
Indeed, ambron's claim that high quality ancient samples are removed from analyses because they're too high quality is one of the most idiotic things I've ever read here, and that's really saying something.
You won't learn anything from ambron about ancient DNA or the ancestry of the Wielbark Goths. He might be an expert in something, but he's not an expert in genetics.
@ Romulus
''It's funny the Wikipedia article titled "Origin of the Goths" starts out "Concerning the origin of the Goths before the 3rd century, there is no consensus among scholars." and cites two sources published fairly recently''
Yeah, citing some 'neo-scholars' who just decided amongst themselves that migrations, peoples and cultures aren't a thing. I guess they didnt bank on scientific evidence catching up with their theoretical / philosophical platforms.
I used to edit Wiki ~ 20 years ago, might have to make a comeback and set some records straight
@ JS- thanks
David, Lucas
David is an eminent specialist in genetics. But we are not talking about people here, we are talking about scientific problems.
Regardless of the words I used to describe the problem, the crux of the problem is that samples contaminated with modern DNA are excluded from further analysis at the initial stage of testing. So the Slavic genetic variants found in the Wielbark population cannot be contamination with modern DNA, as David suggests. It is worth noting that David does not address the crux of the problem, but clings to words. And in the absence of factual arguments, David employs eristics, depreciating the competence of the adversary.
I am indeed not an expert in genetics (although genetics is not foreign to me, as I am a pharmacologist), but I am entitled to my opinion and to express it in a public forum. And of course, I do not change my opinion: the Slavic haplotypes of the Y chromosome in the Wielbark Goths are further evidence for the mixed Scandinavian-Slavic character of the Wielbark population.
My opinion can of course be ignored, as David advises.
Steppe said...@ ph2ter
"Thank you, it's all good and IA IA_Prescythian_Mezocsat?"
Of Mezoscat samples only I18211 shows some Scythian:
HUN_EIA_Prescythian_Mezocsat__I18211_-775 pen=0.01
-------------- ANCESTRY BREAKDOWN: -------------
25.870% CZE_LBA_Knoviz__I15955_-1050
13.131% CZE_MBA_Tumulus__I16112_-1375
12.430% HRV_Bezdanjaca_BA__I18087_-1500
12.284% CZE_LBA_Knoviz__I13783_-1050
10.260% DEU_Tollense_BA__WEZ57_-1200
4.503% CZE_LBA_Knoviz__I13788_-1050
3.976% KAZ_Zevakinskiy_LBA__I3977_-935
3.566% CZE_LBA_Knoviz__I16549_-1050
3.543% HUN_LBA__I20773_-1250
3.363% Saka_Kazakh_steppe_o2__DA19_-775
2.521% ARM_Black_Fortress_LBA__I18247_-1175
2.258% DEU_Tollense_BA__WEZ61_-1200
1.732% KAZ_Aktogai_MLBA__I10140_-1300
0.563% KAZ_Dali_MLBA__I1931_-1180
------------------------------------------------
Fit error: 0.01195702567670328
And this is for IR1:
HUN_Prescythian_IA__IR1_-905
-------------- ANCESTRY BREAKDOWN: -------------
21.254% RUS_Tagar__DA8_-850
12.562% SRB_IA__I16814_-953
12.300% MDA_Cimmerian__cim358_-875
11.960% ITA_Proto-Villanovan__RMPR1_-885
10.778% RUS_Krasnoyarsk_MLBA__I1852_-1345
9.981% KAZ_Lisakovskiy_MLBA_Alakul__I6800_-1465
6.111% ARM_Black_Fortress_LBA__I18247_-1175
5.740% CZE_LBA_Knoviz__I13787_-1050
4.498% ARM_MBA__RISE423_-1558
2.558% DEU_Tollense_BA__WEZ40_-1200
1.720% HRV_MBA_LBA__I18719_-1250
0.537% ARM_Dzori_Gekh_LBA__I16120_-1440
------------------------------------------------
Fit error: 0.01618750696688211
@ Ph2ter
Thanks you are an expert in Calculators
New paper on a technical discussion of qpAdm for anyone interested:
https://www.biorxiv.org/content/10.1101/2023.11.13.566841v1
From what I skimmed of it the main points seem to be:
-qpAdm performs well under population divergence on the order of SW Asian Iron-Bronze Age and older, converging on a small set of plausible candidate models, which frequently includes the true sources but is not as good at giving you the true source unless the genomes you are dealing with are very high coverage and all other assumptions are met (such as not having an isolation-by-distance cline, left to right pop gene flow etc)
-QpAdm seems unaffected by complex admixture histories in the sources and if anything seems to improve its performance on average, also not affecting admixture weight estimation or causing other biases if the true source population is identified/selected for the model.
-Generations past since admixture don't seem to influence qpAdm much either and
-If you have high coverage genomes p-values generally reflect reality and it's not a bad idea to use ranking then.
Romulus said...
"The Goths are exactly who the Roman writers said they were."
There's a problem there because the Roman writers have them pretty much all over the place. Jordanes uses the name "Getae" which looks like the name refer to other folks who were in contact with the Greeks.
What would be interesting is how the aDNA we have can confirm the migration to or from the area north of the Black Sea mentioned so often in both Roman and Old Norse materials. Is there genetic evidence of "Goths" in Iron Age Ukraine?
What's the story with the genetics they've found so far from Chernyakhov culture?
https://en.wikipedia.org/wiki/Origin_stories_of_the_Goths
@ steppe
Don’t know what your obsession with Calculators is, they can produce over fitted results, although they’re okay for an initial screen.
Also, it doesn’t look good when you’re blindly arguing topics you haven’t bothered to check or familiarise yourself.
@LivoniaG
What's the story with the genetics they've found so far from Chernyakhov culture?
They look mixed, but largely Germanic.
Distance to: Ukraine_Chernyakhiv.SG:MJ-19_noUDG.SG
0.03739921 Slovakian
0.04221199 Polish_Silesian
0.04362945 Czech
0.04468093 Polish
0.04504425 Ukrainian_Zakarpattia
0.04560545 Sorb_Niederlausitz
0.04763502 German_Hamburg
0.04764958 Ukrainian_Rivne
0.04766981 Hungarian
0.04840504 Ukrainian_Dnipro
@ Davidski:
Can you elaborate here? I am not sure how the above numbers (if understand them correctly) square with your statement.
Davidski said... "They look mixed, but largely Germanic."
David - please forgive my ignorance --
Are these numbers that you posted showing you that Chernyakhov is largely Germanic? Sorry if this is a stupid question.
"Distance to: Ukraine_Chernyakhiv.SG:MJ-19_noUDG.SG
0.03739921 Slovakian
0.04221199 Polish_Silesian
0.04362945 Czech
0.04468093 Polish
0.04504425 Ukrainian_Zakarpattia
0.04560545 Sorb_Niederlausitz
0.04763502 German_Hamburg
0.04764958 Ukrainian_Rivne
0.04766981 Hungarian
0.04840504 Ukrainian_Dnipro
Many thanks for the analysis, ph2ter. Re IR1, do I get it right that Krasnoyarsk_MLBA is the same as Yakutia_MLBA or is it just kra001?
@Lucas Slawbuski
It's a distance analysis comparing the Chernyakhiv Goth to modern populations.
The reason that West Slavic populations are at the top of the list is because these modern groups have a similarly mixed background to the Goth. That is, Slavic/Germanic.
However, the Goth is more Germanic than the West Slavs. We can tell because the distances are actually fairly high, which means that there are no really close matches on the list.
Also, a couple of populations from Germany appear on the list (Sorb_Niederlausitz, German_Hamburg) as well as modern Hungarians, and all of these groups are largely Germanic/Slavic in terms of genetics.
Modern DNA is very deceptive. So much so that there are still people arguing online that Poles are basically pure Slavs, and they probably will continue to do this for many years.
@LivoniaG
Are these numbers that you posted showing you that Chernyakhov is largely Germanic? Sorry if this is a stupid question.
Yes, this may seem counter intuitive, but they actually do show this.
Also, in European PCA the Chernyakhiv Goth clusters very strongly with modern North Germans.
I see. Thanks. Sorb would expect to be Slavic though and Hamburg was close to the edge of Slavic settlement (if memory serves Slavs burnt it down once), that is not sure that’s a great proxy for north Germans (I kind of think of North Germans as close to Danes or Frisians/Dutch). It’s also interesting that Slovaks for whom Slavs were presumably an eponymous population would be so far ahead of the others on this list (though maybe you .037 isn’t a great fit in any event).
More to the point what would you consider a “good fit” in the sense of “close” distances in the above scale?
@Lucas Slawbuski
Slovaks overlap significantly in terms of genetics with eastern Germans and Austrians, and that's because all of these groups are of mixed Slavic/Germanic origin.
If the Chernyakhiv Goth wasn't mixed, we would probably see some matches of less than 0.03.
The significantly Germanic character of the Chernyakhiv Goths was discussed in the paper that these samples were published with, and I'm pretty sure that you'll eventually see them clustering with Scandinavians in IBD graphs.
David
Can you show us the distances of the Masłomęcz outlier in the same way?
Looks much more northerly and basically East Slavic, probably because of the significant Baltic ancestry.
Distance to: Poland_Maslomecz_IA_o:PCA0103
0.03023034 Russian_Smolensk
0.03047729 Polish_Kashubian
0.03135848 Belarusian
0.03236435 Polish
0.03280527 Russian_Kaluga
0.03295912 Russian_Voronez
0.03314053 Ukrainian_Sumy
0.03443971 Ukrainian_Rivne
0.03544554 Russian_Orel
0.03581461 Russian_Kursk
0.03589781 Russian_Pskov
0.03622715 Ukrainian_Zhytomyr
0.03694963 Ukrainian_Chernihiv
0.03757315 Lithuanian_PA
Well, these mixed Gothic samples from eastern Poland and deep in Ukraine are probably quite telling about where the earliest Slavs lived.
The Goth from Poland has Baltic-like ancestry, while the Goth from Ukraine is more Slavic-like.
It is obvious that Chernyakhiv sample is half Germanic:
UKR_Chernyakhiv_Legedzine__MJ19_400
-------------- ANCESTRY BREAKDOWN: -------------
37.763% POL_IA_Wielbark__PCA0060_115
32.464% HRV_Osijek_Balto-Slavic_o__R3657_280
8.997% POL_IA_Wielbark__PCA0476_200
8.112% POL_IA_Wielbark__PCA0040_160
6.338% ARM_Beniamin__R11651_71
3.065% KAZ_Otyrar_Antiquity__KNT005_319
2.152% Baltic_LTU_Bailuliai__R10840_339
0.963% POL_IA_Wielbark__PCA0065_200
0.146% DEU_Mecklenburg-Vorpommern_Haeven__HVN007_300
------------------------------------------------
Fit error: 0.026043383142592566
Here are the distances of these samples to modern populations:
Distance to: HRV_Osijek_Balto-Slavic_o__R3657_280
0.02842126 Russian_Pskov
0.03106225 Russian_Novgorod_s
0.03132928 Lithuanian_VA
Distance to: POL_IA_Wielbark__PCA0065_200
0.04407501 Norwegian_East
0.04681857 German_Mecklenburg_s
0.04716448 German_Farther_Pomerania_s
Distance to: POL_IA_Wielbark__PCA0476_200
0.04560323 German_Mecklenburg_sx
0.04746873 German_Hamburg
0.04778762 German_Schleswig-Holstein_s
Distance to: POL_IA_Wielbark__PCA0040_160
0.02958899 Swedish_Central_South
0.02978729 Swedish_South
0.02987668 Swedish_Ostergotland_s
Distance to: POL_IA_Wielbark__PCA0060_115
0.03525276 Swedish_Central_North
0.03579073 German_Mecklenburg_sx
0.03632005 Swedish_Varmland_s
@Queequeg said...
Many thanks for the analysis, ph2ter. Re IR1, do I get it right that Krasnoyarsk_MLBA is the same as Yakutia_MLBA or is it just kra001?
Only kra001 is autosomally Yakutia_MLBA (Nganassan like).
Krasnoyarsk_MLBA is a Steppe population.
Yeah, these mixed Goths are first generation Germanic/Balto-Slavic mixes.
So they actually catch the moment when the present-day Eastern European gene pool was forming.
That's the power of ancient DNA!
David
The Slavs were supposedly more Baltic-like in their homeland.
@ambron
Slavs were probably never exactly like Balts. They seem to have been always more southern right from their inception.
But yeah, they were very Baltic-like and they still are.
@ Rob
everyone can make mistakes and back then Anthrogenica (2017) presented it like this that D112 had an East Asian input, of course we are getting more and more new genetic samples but there was contact between the Hallstatt zone and the Pontic Steppe!
https://www.google.com/url?q=https://www.researchgate.net/publication/347323199_Chronological_schemes_of_the_Late_Hallstatt_period_HaD_in_Central_Europe_new_opportuni-ties_for_the_synchronization_and_refinement_of_dates&sa=U&ved=2ahUKEwjgp42w0sqCAxURhP0HHftsB p8QFnoECCMQAQ&usg=AOvVaw245gj-akMmzOXjWhbqvBH_
https://www.google.com/search?q=halstatt+contact+to+scythian&client=safari&sca_esv=583283884&hl=de-de&sxsrf=AM9HkKlnDQ2ZY05ZqNphLd9XIypMU4V2ug%3A1700210946910&ei=AilXZYyYN5-U9u8P--i7oAw&udm=&oq=halstatt+contact+to+scythian&gs_lp=EhNtb2JpbGUtZ3dzLXdpei1zZXJwIhxoYWxzdGF0dCBjb250YWN0IHRvIHNjeXRoaWFuMgcQIRigARgKMgcQIRigARgKSKCKAVC9EliAiAFwCXgBkAEAmAHDAaABkyWqAQQwLjM0uAEDyAEA-AEBqAIPwgIHECMYsAMYJ8ICChAAGEcY1gQYsAPCAg0QIxiABBiKBRjqAhgnwgIHECMY6gIYJ8ICDRAuGIAEGIoFGOoCGCfCAgoQIxiABBiKBRgnwgIKEC4YgAQYigUYJ8ICERAuGIAEGLEDGIMBGMcBGNEDwgILEC4YgAQYsQMYgwHCAgsQABiABBixAxiDAcICDhAAGIAEGIoFGLEDGIMBwgIEECMYJ8ICChAuGIAEGIoFGEPCAgUQLhiABMICChAAGIAEGIoFGEPCAg4QLhiABBiKBRixAxiDAcICEBAuGIAEGBQYhwIYsQMYgwHCAhYQLhgUGIMBGK8BGMcBGIcCGLEDGIAEwgIFEAAYgATCAgoQABiABBgUGIcCwgIKEC4YFBiHAhiABMICDBAuGIAEGIoFGAoYQ8ICDRAuGIAEGIoFGLEDGEPCAggQABiABBjJA8ICCxAAGIAEGJIDGLgEwgIIEC4YgAQYsQPCAg4QLhiABBixAxiDARjUAsICBxAAGIAEGArCAgcQIxixAhgnwgIKEC4YsQMYgAQYCsICBxAjGLACGCfCAgoQLhgNGLEDGIAEwgIKEAAYgAQYDRixA8ICBxAAGIAEGA3CAgoQABiABBjLARgKwgIKEC4YywEYgAQYCsICCRAAGIAEGA0YCsICBhAAGB4YDcICCRAAGBYYHhjxBMICBhAAGBYYHsICCRAAGIAEGA0YE8ICCBAAGBYYHhgTwgIKEAAYCBgeGA0YE8ICDBAAGAgYHhgNGA8YE8ICBBAhGBXCAggQIRgWGB4YHcICCxAhGBYYHhjxBBgdwgIFECEYoAHiAwQYACBBiAYBkAYJ&sclient=mobile-gws-wiz-serp
David
I propose to note that the distance of Chernyakhov to the nearest Polish population is 0.04221199, while Masłomęcz is 0.03047729 (a cosmic difference). In other words: the Poles share much more origin with Masłomęcz than with Chernyakhov.
@ambron
You can't read the distance scores so literally.
Poles are much closer to the Polish Goth because the Baltic input shifts her east and makes her look less Germanic.
@Davidski
Is Av2 omitted from your ancient spreadsheet because it is a relative of Av1?
Av2 is less mixed than Av1 and is a more representative of Slavic EMA populations than Av1.
David
I know this. It is important that others remember this and that this rule works both ways, as for example in this case:
Distance to: POL_IA_Wielbark__PCA0065_200
0.04407501 Norwegian_East
0.04681857 German_Mecklenburg_s
0.04716448 German_Farther_Pomerania_s
The huge distance will show that something is missing here. What is missing? Maybe Balto-Slavic genetic variants missing?
@ph2ter
Probably, because I remove all close relatives.
@ambron
As per the discussion above, there's something strange about that sample and another one like it.
It seems they might have some sort of far eastern input from the steppe or even something Uralic.
Or they're just really poor sequences.
David
The coverage cannot be very poor, because the distance to modern populations is not bad:
Target: Poland_Kowalewko_IA:PCA0065
Distance: 3.0847% / 0.03084709
56.2 Polish_Kashubian
38.2 Irish
2.6 Biaka
2.2 Brahmin_Uttar_Pradesh
0.8 Saharawi
But we can actually see some exotic signals.
@ambron
The fact that we're seeing these African signals means there's a serious coverage and damage problem.
2.6 Biaka
0.8 Saharawi
@ steppe - all good old friend
gamerz_j: "New paper on a technical discussion of qpAdm for anyone interested: https://www.biorxiv.org/content/10.1101/2023.11.13.566841v1"
thanks for the summary on the points you thought of note. (no doubt there are many more points within too). I found this paper incredibly dense!
While I didn't take in much I thought it was surprising to me to see them say this at the end:
"A surprising finding from the complex Eurasian demographic simulations was when performing the qpAdm rotation analysis with f2-statistics computed from the whole-genome branch lengths we obtained a 100% true positive rate and a 0% false positive rate with a P-value threshold of 0.001 (SI Figure S6). This outcome underscores the remarkable potential inherent in the principles underlying qpAdm, while also highlighting the constraints imposed by data scale. In light of this, we suggest there is room for significant enhancements in the qpAdm protocol from methods that can leverage more accurate estimations of f2-statistics. Possible avenues for this improvement could be developing innovative techniques for extracting information from ancestral recombination graphs (ARG) within the context of aDNA, or conditioning on the site frequency spectrum (SFS) for the enrichment of rare alleles. As such, it is clear from these results that further improvements would make qpAdm a powerful tool for accurately reconstructing the genetic histories of populations under the most complex scenarios."
Where I've always thought that whole-genomes seemed to be important was for reconstructing the ground truth of the site frequency spectrum (which you don't really do in capture data which is based on ancestral SNP polymorphisms) and so these models that understand the dynamics of population growth. (As in Marchi 2022 "The genomic origins of the world’s first farmers", where they argued effectively as I understand it that the European HG population had a large bottleneck during the LGM but was actually part of a recently fairly large population while the Early Farmers had avoided that but recently undergone admixture and a sharp bottleneck prior to expanding through Europe. Even if there are questions marks about the deeper population model there).
What you get from whole-genomes, I thought, is rare variants that are not even shared widely within a population, let alone all human populations. But the level of variation of allows, even if not the specific sharing, allows you to reconstruction population size history.
It's surprising then to me that these variants which are not shared widely, assuming I've understood this correctly, are actually important for qpAdm and presumably qpGraph? It would be interesting if they'd said more about the reasons about why they thought this was the case. Why do rare variants contribute to the f4 ABBA vs BABA statistics which are supposed to drive qpAdm?
But there are also better, interesting samples - small distance to contemporary populations, little noise:
Target: Poland_Kowalewko_IA:PCA0063
Distance: 2.2381% / 0.02238149
48.0 Danish
22.0 Latvian
19.2 Irish
6.2 Basque_Spanish
4.2 English_Cornwall
0.2 Biaka
0.2 Koinanbe
Target: Poland_Kowalewko_IA:PCA0027
Distance: 2.5998% / 0.02599809
37.0 Latvian
27.4 Norwegian
21.4 Icelandic
11.8 Basque_French
1.6 Igorot
0.8 Ju_hoan_North
By entering into a discussion with Ambron, you ennoble this fanatic.
I would like to remind you that a geneticist from Warsaw claims that the Wielbark samples, especially from Masłomęcz, are most likely mixed with later ones and confused.
@ambron
Those aren't distances, they're overfitted ancestry proportions.
When you see Basque, Latvian and other unrealistic sources appear in the same model, it means the model is statistically overfitted.
The Latvian is canceled out by the Basque, so there's nothing Baltic or Slavic there.
You'd be able to tell if these Wielbark Goths had significant Baltic ancestry, especially at 20-40%, because they'd cluster well east of the Scandinavian cluster.
But they don't.
@galadhorn
Most of the Wielbark samples are of very poor quality, but they look genuine to me in terms of genetic structure and also C14 dates.
Even the singleton outlier looks genuine, because it's not unrealistic to see a Goth/Balt mix at that time in eastern Poland.
The problem here is that we're trying to work out very fine scale ancestry using low quality sequences.
So the interpretation of the results can be twisted by people who don't understand them fully and want to see something that isn't there.
It's frustrating, but eventually there will be more samples and also much higher quality samples, as well as new results based on haplotypes and IBD. So in the end there won't be anything to twist, and ambron will either have to accept reality or disappear from these discussions.
@All
Here are the actual distances for the samples that ambron posted ancestry proportions.
Distance to: Poland_Kowalewko_IA:PCA0063
0.02600474 Danish
0.02692912 Swedish
0.02934648 Norwegian
0.02944747 German_Hamburg
0.02990236 Icelandic
0.03359235 Dutch
0.03388140 Orcadian
0.03506714 Scottish
0.03546483 Welsh
0.03621106 Irish
Distance to: Poland_Kowalewko_IA:PCA0027
0.03292472 Swedish
0.03899474 German_Hamburg
0.04039804 Norwegian
0.04144449 Danish
0.04284346 Icelandic
0.04558798 Polish_Kashubian
0.04560316 Czech
0.04627336 German_East
0.04713319 Slovakian
0.04714277 Polish
Distance to: Wielbark_culture_Kowalewko_o:PCA0002
0.03520301 Ukrainian_Volyn
0.03809343 Cossack_Ukrainian_Dnipropetrovsk
0.03881902 Russian_Lipetsk
0.03902537 Ukrainian_Donetsk
0.03950398 Ukrainian
0.04016299 Ukrainian_Crimea
0.04037858 Russian_Smolensk
0.04041922 Ukrainian_Sumy
0.04141225 Ukrainian_Poltava
0.04186307 Russian_Oryol
0.04186389 Belarusian
0.04193451 Polish
Distance to: Wielbark_culture_Maslomecz_o:PCA0103
0.02490921 Polish_Kashubian:Kashubian1
0.02646948 Polish_Kashubian
0.02848002 Russian_Smolensk:RUS_Smol339
0.02852873 Russian_Kursk:Rkuch-05
0.02871251 Polish:Polish8
0.02912088 Polish:Polish32
0.02940583 Ukrainian_Sumy:EG600077
0.03023034 Russian_Smolensk
0.03051155 Ukrainian_Chernihiv:EG600056
0.03122534 Russian_Orel:Rorl-155
0.03135848 Belarusian
0.03146193 Belarusian:Belarusian2
0.03157803 Polish:Polish24
0.03160156 Russian_Orel:russianOrjol1
0.03161597 Polish:Polish11
0.03201117 Russian_Smolensk:RUS_Smol304
0.03229449 Russian_Smolensk:RUS_Smol350
0.03236435 Polish
0.03236719 Russian_Smolensk:RUS_Smol345
0.03255961 Polish:Polish40
0.03280527 Russian_Kaluga
0.03295912 Russian_Voronez
0.03297586 Polish:Polish37
0.03310648 Ukrainian_Sumy:EG600084
0.03311215 Belarusian:Belarusian12
PCA0002 Y-DNA - R1a-M458-CTS11962
David
We have just talked about the fact that we need to draw conclusions from modern distances carefully and symmetrically. Today's Scandinavians and Germans have a lot of Balto-Slavic admixture that the native Iron Age Germanics did not have:
Baltic BA:
German East 27.6
Swedish 14
German 13
Danish 9.6
DNK Jutland IA VK582 0.0
https://slavicorigins.blogspot.com/2021/05/baltic-ba-ancestry-in-wielbark-culture.html
David
Gladhorn doesn't know anything about genetics, so all he can do is make me look crazy. A discussion is a presentation of different views. Unanimity is not discussion. Not everyone with a different view is crazy. According to genetic and linguistic data, I see the Slavic homeland in the area of historic Galicia, but I respect the opinion of anyone who places it somewhere else on the map of Europe.
Gladhorn should be grateful to me, because if it weren't for my provocation, you would probably never have dealt with the Wielbark and Trzciniec genomes.
@ambron
In order to prove that the Wielbark population is significantly mixed with Balts and/or Slavs, you need to show that relatively high quality and contamination free Wielbark genomes consistently cluster between Scandinavians and Balts.
So, for instance, if the Wielbark samples by and large resembled Poland_Maslomecz_IA_o:PCA0103 and/or Ukraine_Chernyakhiv.SG:MJ-19_noUDG.SG in terms of genetic affinities then we could say without any doubt that you have a point.
But you don't have a point, because claiming that modern Scandinavians are part Balto-Slavic, and so the Wielbark population's strong Scandinavian genetic character just means that they're part Balto-Slavic is not a convincing argument.
That's because it's not true that Scandinavians by and large have significant Balto-Slavic admixture. Only the more easterly Scandinavian populations do.
And, in fact, in terms of basic Northwest/Southeast affinities the Wielbark genomes, when grouped into populations, actually come out more Scandinavian than Viking Age Scandinavians.
https://blogger.googleusercontent.com/img/b/R29vZ2xl/AVvXsEjVOAGHspJcbOFh5gbhwYjKbqie5ys6nHjeNhWRSXESjTD1kMNf5sB-NsMXnNORnu9VC4190FyV7Stt38jM7WJbH1MzxArtxpcVhgU62bgozGeGiGp31JzI3_laVzQ0eaYkwdxN-9mq2BKdE14AmeM60WoKBCZ1Js_9Zc7QAyHqc113Q_sJd3zX_ZpryeYN/s1279/Poland_Wielbark_IA_f3_graph.png
David
What explanation do you propose for the location of Weklice according to f3 (Cameroon, Estonia) between Nimcza and Ląd?
@ambron
It's just one stat. You have to combine stats to get a more accurate picture because there are often overlaps between such similar groups.
There's no way you can say Weklice_IA is part Balto-Slavic based on this stat.
f3(X,Ireland_Megalithic,Cameroon_SMA)
Poland_Maslomecz_IA 0.219816
Poland_Weklice_IA 0.219501
Denmark_Viking 0.2192
Poland_Kowalewko_IA 0.219176
Poland_Ostrow_Lednicki_MA 0.218916
Norway_Viking 0.218854
Poland_Pruszcz_Gdanski_IA 0.218684
Sweden_Viking 0.218626
Denmark_EarlyViking 0.218529
England_Viking 0.218308
Russia_Viking 0.217999
Poland_Viking 0.217914
Poland_Plonsk_MA 0.217756
Poland_Lad_MA 0.217719
Poland_Legowo_MA 0.21765
Poland_Niemcza_MA 0.217001
Poland_Trzciniec_Culture 0.216551
David
But it can be said that Weklice shares approximately the same number of genetic variants with the Baltic BA as Niemcza and Ląd.
The Chernyakhov sample also shows us that we cannot look for a Slavic homeland too far east of Poland, because modern Poles do not have any Central Asian admixture:
Target: UKR_Chernyakhiv_Legedzine:MJ19
Distance: 2.3159% / 0.02315851 | R3P
69.0 Polish
21.8 Swedish
9.2 Tajik_Shugnan
In some new Polish paper will be plenty Wielbark but also Przeworsk genomes which will confirm what David tells us. Just wait till next year. Ambron will hide then;)
I have something interesting for you from Krzysztof Narog from "Genealogia Genetyczna" on Facebook:
Quote:
"Visualization of the distribution of halogroups in Poland. For greater readability, I marked the dominant R1a and R1b with outlines
I developed it based on data from: https://hras.yseq.net/, which means it is based on ancestral data - it is not exactly the modern distribution of Y."
See picture: https://genarchivist.com/attachment.php?aid=260
and see R1a! It looks like it was entering today's Poland from the area of today's Ukraine (Volhynia) in the past.
@Davidski
Could you run those samples https://ufile.io/zt7keyp2
(I've put only the highest coverage ones although they're all relatively low) from:
https://doi.org/10.1016/j.cub.2023.09.055
https://www.researchgate.net/publication/374801698_Ancient_genomes_reveal_millet_farming-related_demic_diffusion_from_the_Yellow_River_into_southwest_China
Thanks!
@galadhorn
Based on these sorts of maps some people are still arguing that R1b is from Iberia and R1a from South Asia.
@crashdoc
These sequences and/or the genotyping are terrible, with high levels of African ancestry showing up.
@Davidski
Ok, in the article it's mentionned that they had to do some trimming to get them cleaned-up, so I guess we'll have to wait for the official genotypes.
I had a look at the Weklice 'Goths' with qpAdm, they indeed just come out as one way admixutre from Denmark_IA (Sweden_IA also works, but order of magnitute lower tail prob).
No Latvia_BA
Poland_Weklice2
Denmark_IA.SG
Latvia_BA
best coefficients: 1.084 -0.084
Tail prob
0.365966
0.369023
right pops:
Cameroon_SMA
Morocco_Iberomaurusian
China_Tianyuan
Russia_MA1_HG.SG
Italy_North_Villabruna_HG
Serbia_IronGates_Mesolithic
Turkey_Epipaleolithic
Iran_GanjDareh_N
Turkey_N
Ethiopia_4500BP
Czech_Vestonice
Russia_DevilsCave_N.SG
Russia_Kostenki14
Germany_Mesolithic
Sweden_Ansarve_Megalithic.SG
Sweden_BAC.SG
Sweden_Motala_HG
Poland_GlobularAmphora
Estonia_CordedWare.SG
Denmark_Djursland_SingleGraveCulture.SG
Germany_Tollense
Russia_Bolshoy
Russia_IA_Ingria.SG
Rob
The upcoming work with a new collection of samples from Weklice, the results of which were initially pre-presented at the conference, clearly shows using IBS segments and f3 and f4 statistics that Weklice was a mixed Scandinavian-Slavic population.
@ambron
IBD segments, not IBS segments.
Who's involved in this new Weklice study? Stolarek again?
Greetings, dear Davidski! I came across on the Internet the simulated coordinates of Caucasian hunters who participated in the ethnogenesis of people, like eneolitic progress, and according to your calculator it turned out that in terms of distance he is further from the Eastern European hunters than the same hunters from the Kotias cave, despite the fact that Kotias is located south of the Caucasus mountains, and progress is the northern Caucasus, it seems to me that the person who modeled these coordinates completely removed everything related to EHG, including ANE .
Is it possible to assume that the Caucasian hunter-gatherers of progress had fewer genes associated with ANE and EHG?
Distance to: Progress_CHG_Simulated
0.37347121 RUS_Samara_HG:I0124
0.38436417 RUS_Sidelkino_HG:Sidelkino
0.39016294 RUS_Karelia_HG:UzOO77
0.39246257 RUS_Karelia_HG:I0061
0.40740622 RUS_Tyumen_HG:I1960
0.40870510 RUS_AfontovaGora3:AfontovaGora3
0.40998461 RUS_Karelia_HG:I0211
Distance to: GEO_CHG:KK1
0.35614292 RUS_Samara_HG:I0124
0.36726873 RUS_Sidelkino_HG:Sidelkino
0.37130728 RUS_Karelia_HG:UzOO77
0.37361704 RUS_Karelia_HG:I0061
0.38349246 RUS_Tyumen_HG:I1960
0.38776927 RUS_AfontovaGora3:AfontovaGora3
0.39547366 RUS_Karelia_HG:I0211
Distance to: CHG_Mesolitic:NEO281
0.36086863 RUS_Samara_HG:I0124
0.37178463 RUS_Sidelkino_HG:Sidelkino
0.37433235 RUS_Karelia_HG:UzOO77
0.37843292 RUS_Karelia_HG:I0061
0.38503753 RUS_Tyumen_HG:I1960
0.38710385 RUS_AfontovaGora3:AfontovaGora3
0.39983165 RUS_Karelia_HG:I0211
here's another
Target: CHG_Mesolitic:NEO281
Distance: 14.2141% / 0.14214057
75.2 Iran_Neolithic
14.8 Early_European_Farmer
10.0 Eastern_Hunter-Gatherer
Target: Progress_CHG_Simulated
Distance: 14.4227% / 0.14422706
75.0 Iran_Neolithic
19.4 Early_European_Farmer
5.6 Eastern_Hunter-Gatherer
Target: GEO_CHG:KK1
Distance: 15.2229% / 0.15222940
71.4 Iran_Neolithic
16.6 Early_European_Farmer
12.0 Eastern_Hunter-Gatherer
@Арсен
I don't work with simulated coordinates.
Also, like I said in my recent blog post, there was no migration from the Caucasus or from south of the Caucasus into the steppe.
It was a diffusion of alleles from different areas near the Caucasus that took a long time, starting at least 5,500 BCE.
https://eurogenes.blogspot.com/2023/09/the-caucasus-is-semipermeable-genetic.html
You can't really simulate that sort of process with one type of population.
David
In this case, these are IBS segments, not IBD segments, analyzed in pairs.
This is the Łódź team that publishes with Fernandes. The already known genomes from Weklice and all the genomes with the "N" signature come from them - N17, N47, N49, etc.
Post a Comment