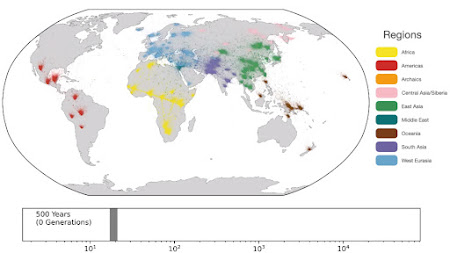
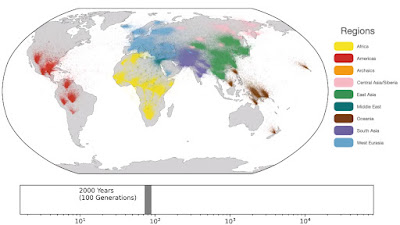
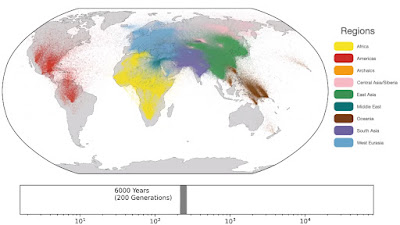
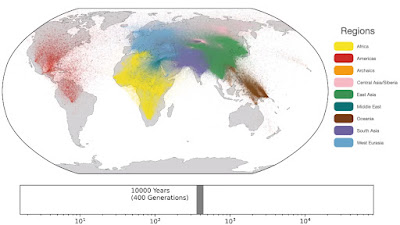
Spatio-temporal dynamics in human history. This movie shows the estimated geographic locations of ancestors of Human Genome Diversity Project, Simons Genome Diversity Project, Neanderthal, Denisovan, and Afanasievo samples over time. Each dot represents an edge in the tree sequence of chromosome 20, where the time and geographic location of the parent and child nodes of the edge have been estimated. The locations of edges at each point in time are plotted along the great circle between the parent and child nodes. Edges are colored by the region of the descendants of the child node. If an ancestral lineage has ancestors in multiple regions, its color is the average of the respective colors of each region.See also... Haplotype-based PCA of West Eurasia and Europe
search this blog
Thursday, February 24, 2022
A unified genealogy of modern and ancient genomes (Wilder Wohns et al. 2022)
Over at Science at this LINK. Broadly speaking, this looks like a more sophisticated version of something that I tried about five years ago (see here).
I wonder if they got the idea from me? Honestly, I wouldn't be surprised if they did. But like I say, their methods are way more advanced.
Keep in mind, however, that for now, their analysis includes 3601 modern genomes and just eight ancient genomes. That's because they can only run super high quality ancient sequences. The ratio of ancient genomes will no doubt rise rapidly over the next few years, and that's when things will get really interesting.
Below are some screen caps from a clip accompanying the paper, freely available here. This is the caption to the movie:
Monday, February 21, 2022
The Pict
KD001 is the first undeniable Pictish sample in my dataset, courtesy of Dulias et al. 2022. Thanks to Altvred for processing the files.
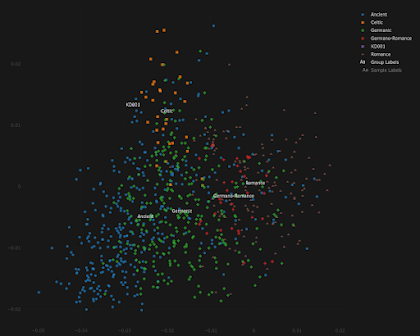
This is how KD001 behaves in my Celtic vs Germanic Principal Component Analysis (PCA). Looks kind of Irish, doesn't he?
To see an interactive version of the plot, paste the coordinates from here into the relevant field here.
See also...
Celtic vs Germanic Europe
Avalon vs Valhalla revisited
When did Celtic languages arrive in Britain?
Labels:
Albion,
ancient DNA,
Britain,
British DNA,
British Isles,
Celtic,
Celtic vs Germanic,
Great Britain,
Indo-European,
Northern Europe,
Northwestern Europe,
Pict,
Pictish,
Picts,
Scotland,
Scottish
Monday, February 14, 2022
Blond hair is only indirectly associated with Anatolian ancestry in Estonia...duh
In a recent paper about complex traits in Europeans, Marnetto et al. found that blond hair and blue eyes showed a relatively high association with ancient Anatolian ancestry.
This is a somewhat curious finding considering that ancient Anatolians weren't particularly blond haired or blued eyed, and that's probably an understatement.
However, the Europeans that Marnetto et al. based their analysis on were Estonians. And in Estonia ancient Anatolian ancestry peaks in the west and north, probably because this is where Estonians have the most Germanic and Finnish ancestry.
Germanic and Finnish populations are somewhat richer in ancient Anatolian ancestry than Estonians, and, unlike ancient Anatolians, they're often exceptionally blond haired and blue eyed.
So it makes sense that, in Estonia at least, ancient Anatolian ancestry is associated with blond hair and blue eyes, but only indirectly so. The more direct link is between Germanic and Finnish ancestry and blond hair and blue eyes.
I feel that Marnetto et al. should've investigated this, and they also should've made it clear that the associations they found won't necessarily be seen in other European countries.
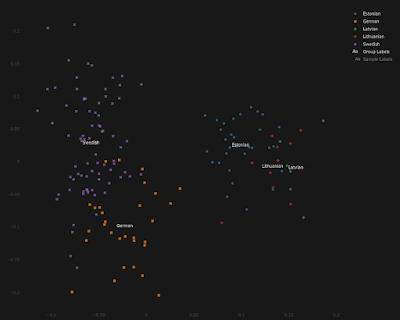
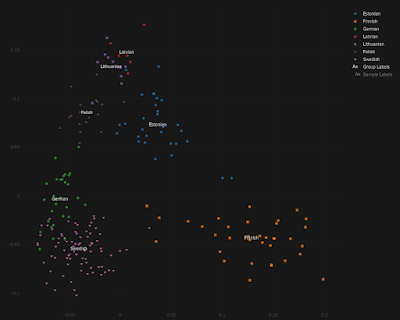
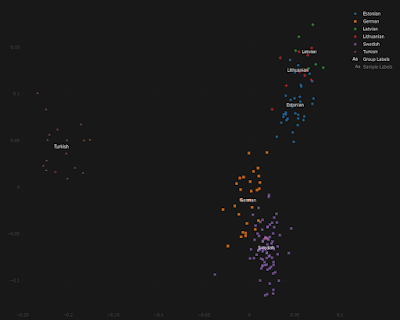
For the doubters out there, and I know there are at least a few of you, below is a series of Principal Component Analyses (PCA) showing how Estonians compare to other populations from around the Baltic Sea, as well as to present-day Turks from central Anatolia.
Note that, by and large, the same Estonians who show more affinity to the Germanic and/or Finnish individuals are also shifted slightly closer to the Turks, and this is because they harbor elevated ancient Anatolian ancestry. The relevant datasheets are available here.
Citation...
Marnetto et al., Ancestral genomic contributions to complex traits in contemporary Europeans, Current Biology (2022), https://doi.org/10.1016/j.cub.2022.01.046
See also...
Ancient ancestry and complex traits in Estonians (Marnetto et al. 2022)
Mainstream media BS: Europeans owe their height to Asian nomads
Thursday, February 10, 2022
Mainstream media BS: Europeans owe their height to Asian nomads
From a recent Daily Mail article by some clown named Sam Tonkin:
Present day Europeans owe their blue eyes to hunter gatherers, their height to Asian nomads and their blonde hair to Anatolian Neolithic farmers, a new study suggests. ... Most of the contemporary European genetic makeup was shaped by movements that occurred in the last 10,000 years when local hunter gatherers mixed with incoming Anatolian farmers — from present-day Turkey — and Asian nomads, or Pontic Steppe pastoralists. The latter originated from what is now parts of Bulgaria, Romania, Moldova, Ukraine, Russia and Kazakhstan.Haha. Bulgaria, Romania, Moldova and Ukraine are European countries. The relevant parts of Russia and Kazakhstan are also located in Europe. Obviously, the author is referring to the Yamnaya herders who lived on the Pontic-Caspian steppe, which is obviously in Eastern Europe. I blame Johannes Krause for this. See also... Matters of (basic) geography Blond hair is only indirectly associated with Anatolian ancestry in Estonia...duh Ancient ancestry and complex traits in Estonians (Marnetto et al. 2022)
Labels:
ancient DNA,
blond,
blonde hair,
Central Asia,
Daily Mail,
Eastern Europe,
height,
Johannes Krause,
media,
North Pontic steppe,
Pontic-Caspian steppe,
propaganda,
Sam Tonkin,
science,
Yamna,
Yamnaya
Wednesday, February 9, 2022
Ancient ancestry and complex traits in Estonians (Marnetto et al. 2022)
Over at Current Biology at this LINK. Here's the summary:
The contemporary European genetic makeup formed in the last 8,000 years when local Western Hunter-Gatherers (WHGs) mixed with incoming Anatolian Neolithic farmers and Pontic Steppe pastoralists. 1–3 This encounter combined genetic variants with distinct evolutionary histories and, together with new environmental challenges faced by the post-Neolithic Europeans, unlocked novel adaptations. 4 Previous studies inferred phenotypes in these source populations, using either a few single loci 5–7 or polygenic scores based on genome-wide association studies, 8–10 and investigated the strength and timing of natural selection on lactase persistence or height, among others. 6,11,12 However, how ancient populations contributed to present-day phenotypic variation is poorly understood. Here, we investigate how the unique tiling of genetic variants inherited from different ancestral components drives the complex traits landscape of contemporary Europeans and quantify selection patterns associated with these components. Using matching individual-level genotype and phenotype data for 27 traits in the Estonian biobank 13 and genotype data directly from the ancient source populations, we quantify the contributions from each ancestry to present-day phenotypic variation in each complex trait. We find substantial differences in ancestry for eye and hair color, body mass index, waist/hip circumferences, and their ratio, height, cholesterol levels, caffeine intake, heart rate, and age at menarche. Furthermore, we find evidence for recent positive selection linked to four of these traits and, in addition, sleep patterns and blood pressure. Our results show that these ancient components were differentiated enough to contribute ancestry-specific signatures to the complex trait variability displayed by contemporary Europeans.This is a fascinating effort, but I'm not taking it too seriously until I see the results reproduced with several cohorts from very different parts of Europe. The reason being is that at least some of the outcomes might be specific to Estonia, and reflective of its own peculiar recent population history. For example, the authors find that among Estonians blond hair and blue eyes show a high association with Anatolian farmer ancestry (see table S4). Now, some people might be surprised by this link between light pigmentation and Near Eastern ancestry. However, I'm not, because I know that quite a few Estonians, especially northwest Estonians, harbor recent north German and/or Scandinavian ancestry. Obviously, north Germans and Scandinavians are some of the blondest haired and lightest eyed people in Europe. But they also have more Anatolian farmer ancestry than Estonians. So it might well be that in Estonia these traits are strongly linked with recent Germanic ancestry rather than ancient Anatolian ancestry. In fact I'm willing to bet that this is indeed the case. I'm also willing to bet that blond hair and blue eyes won't show a strong association with Anatolian farmer ancestry in other European countries, but rather with steppe herder ancestry or even, in some cases, minor Siberian admixture. Citation... Marnetto et al., Ancestral genomic contributions to complex traits in contemporary Europeans, Current Biology (2022), https://doi.org/10.1016/j.cub.2022.01.046 See also... Mainstream media: Europeans owe their height to Asian nomads Blond hair is only indirectly associated with Anatolian ancestry in Estonia...duh
Wednesday, February 2, 2022
The PIE homeland controversy: February 2022 status report
I think we'll see the emergence of two main competing proto-Indo-European (PIE) homeland theories over the next few years:
- a homeland in the Eneolithic North Caucasus, and the spread of Anatolian languages into West Asia with Maykop-related ancestry - a homeland in the North Pontic region, possibly within the Eneolithic Sredny Stog archeological culture, and the spread of Anatolian languages into West Asia via the Balkans.Both theories have support from ancient DNA. Some of it has already been published (for instance, see here). At this point, I can see myself firmly in the North Pontic camp, even if it turns out that North Pontic-related ancestry only made a fleeting impact on Bronze Age Anatolia. After all, there's no direct relationship between genes and languages, so to prove that Anatolian languages came from the North Pontic, there's no need for North Pontic-related ancestry to persist in Anatolia, as long as we have solid evidence that people with this type of ancestry moved there at the right time. In my mind, for now, the Maykop culture provides an excellent explanation for non-Indo-European influences in PIE, and there's no need to make it Indo-European speaking, let alone PIE speaking. See also... The PIE homeland controversy: June 2021 status report
Subscribe to:
Comments (Atom)