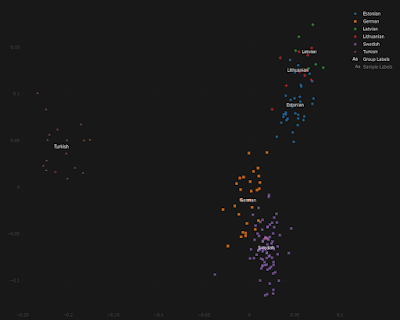
- shows that all Ashkenazim remain genetically linked to a significant degree to other types of Jewish populations, not only paternally but maternally as well - disproves the myth that Cossack rapists were responsible for any of the non-Israelite DNA in Ashkenazim - presents new DNA evidence in favor of a small contribution of Khazarian and Alan converts to Judaism to the Ashkenazic gene pool.That makes good sense based on what I've learned over the years from studying modern and ancient genome-wide Ashkenazic DNA. More information about Kevin's book is available at the Khazaria.com website HERE. See also... My take on the Erfurt Jews
search this blog
Sunday, November 13, 2022
A reappraisal of Ashkenazic maternal ancestry
Kevin Brook, who occasionally comments on this blog, has published a peer-reviewed book titled The Maternal Genetic Lineages of Ashkenazic Jews.
The book focuses on 129 mitochondrial (mtDNA) haplogroups that are found in present-day Ashkenazic Jews, and reveals that these lineages can be traced back to a wide range of places, such as Israel, Italy, Poland, Germany, North Africa, and China.
Ergo, it argues that both Israelites and converts to Judaism from a variety of gentile groups made lasting contributions to the Ashkenazic maternal gene pool. In Kevin's own words, the book also:
Tuesday, November 1, 2022
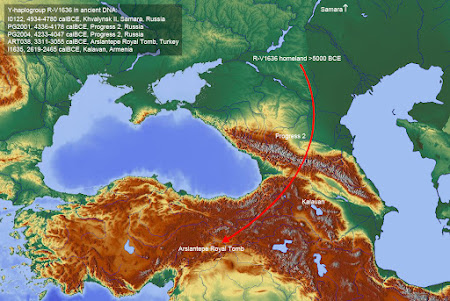
The story of R-V1636
Who wants to bet against this map? Keep in mind that ART038 (~3000 calBCE) remains the oldest sample with the V1636 and R1b Y-chromosome mutations in the West Asian ancient DNA record. Ergo, there's nothing to suggest that V1636 or R1b entered Eastern Europe from West Asia.
See also...
A tantalizing link
How relevant is Arslantepe to the PIE homeland debate?
Thursday, October 27, 2022
The Yassitepe challenge
This is about the only successful qpAdm model that I can find for the pair of Early Bronze Age (EBA) females from Yassitepe, Turkey, using a decent set of outgroups and markers. I wouldn't take it too literally, but it does suggest a potentially significant level of European ancestry, including some steppe ancestry, in these Yassitepe individuals.
TUR_Aegean_Yassitepe_EBA AZE_Caucasus_lowlands_LN 0.565±0.054 ROU_N 0.387±0.041 RUS_Progress_En 0.048±0.022 P-value 0.103248 Full outputIf anyone reading this can find a better, more convincing solution then I'd love to see it. Feel free to share it in the comments below. Obviously, both of the Yassitepe samples are from the recent Lazaridis, Alpaslan-Roodenberg et al. paper. Their EBA dating suggests that they might be relevant to the debate over the origins of Anatolian speakers, such as the Hittites and Luwians. See also... Dear Iosif, about that ~2% The precursor of the Trojans
Thursday, October 13, 2022
The Kura-Araxes people deserve better
When discussing the Kura-Araxes culture and its people it's important to understand these key points:
- there is Eastern European steppe ancestry in Kura-Araxes samples, and if you're not seeing it then you're not looking hard enough - Armenian Kura-Araxes samples are mainly a mixture between three different groups currently best represented in the ancient DNA record by ARM_Areni_C, IRN_Hajji_Firuz_C and RUS_Darkveti-Meshoko_En - ergo, most of the steppe ancestry in the Kura-Araxes population of what is now Armenia must have been mediated via local Chalcolithic groups like ARM_Areni_C - Kura-Araxes samples show Mesopotamian-related ancestry, and this mustn't be ignored.Oh, you don't believe it because you just read a big paper in Science claiming otherwise? Well, the authors of that paper, Lazaridis, Alpaslan-Roodenberg et al., used distal mixture models to study the ancestry of their Kura-Araxes samples, and such models can miss important details. Consider these three proximate mixture models for a relatively high quality and very homogenous Kura-Araxes sample set from the aforementioned paper. They were done with the qpAdm software
ARM_Kura-Araxes_Berkaber ARM_Areni_C 0.239±0.068 IRN_Hajji_Firuz_C 0.379±0.068 RUS_Darkveti-Meshoko_En 0.382±0.054 P-value 0.285122 (Pass) Full output ARM_Kura-Araxes_Berkaber IRN_Hajji_Firuz_C 0.569±0.051 RUS_Darkveti-Meshoko_En 0.363±0.058 RUS_Progress_En 0.068±0.020 P-value 0.20306 (Pass) Full output ARM_Kura-Araxes_Berkaber IRN_Hajji_Firuz_C 0.531±0.060 RUS_Darkveti-Meshoko_En 0.469±0.060 P-value 0.0132579 (Fail) Full outputSome caveats apply. For instance, the pass threshold (P-value ≥0.05) is arbitrary. But the point is that the models look much better with steppe-related and steppe reference populations (ARM_Areni_C and RUS_Progress_En, respectively). Moreover, the unique and vital Darkveti-Meshoko population is represented by just one individual. I also have the genotypes of his brother and sister, but relatives aren't allowed in these sorts of tests. Including a singleton in the analysis means that I can't use the inbreed: YES option, which apparently can be a bad thing. Nevertheless, these models do look very solid. Indeed, I can also model ARM_Kura-Araxes_Berkaber as practically 100% RUS_Maykop_Novosvobodnaya, perhaps with some excess ARM_Areni_C-related input.
ARM_Kura-Araxes_Berkaber ARM_Areni_C 0.094±0.087 RUS_Maykop_Novosvobodnaya 0.906±0.087 P-value 0.284259 (Pass) Full ouputThis makes good sense, because RUS_Maykop_Novosvobodnaya can also be modeled solidly as a mixture between IRN_Hajji_Firuz_C, RUS_Darkveti-Meshoko_En and RUS_Progress_En.
RUS_Maykop_Novosvobodnaya IRN_Hajji_Firuz_C 0.614±0.056 RUS_Darkveti-Meshoko_En 0.307±0.064 RUS_Progress_En 0.080±0.022 P-value 0.141468 (Pass) Full outputI don't know whether the genetic relationship between ARM_Kura-Araxes_Berkaber and RUS_Maykop_Novosvobodnaya shown in my model is due to Maykop ancestry in the former. It might just be a coincidence in the sense that the same or similar processes led to the formation of both groups. Feel free to let me know your thoughts about that in the comments. The fact that the Kura-Araxes people harbored steppe ancestry might be very important in the debate over the location of the so called Indo-Anatolian homeland. For instance, it's possible that the proto-Anatolian language spread from the North Caucasus into Anatolia via the Kura-Araxes culture. But, admittedly, such a solution doesn't have strong support from historical linguistics data, which suggest that the Indo-Anatolian homeland was located in what is now Ukraine and that Anatolian speakers entered West Asia via the Balkans:
Indo-European cereal terminology suggests a Northwest Pontic homeland for the core Indo-European languagesSee also... R-V1636: Eneolithic steppe > Kura-Araxes? Dear Iosif...Yamnaya But Iosif, what about the Phrygians?
Thursday, October 6, 2022
Balto-Slavs and Sarmatians in the Battle of Himera
G25 coordinates for most of the samples from the recent Reitsema et al. paper are available in a text file here. They're also in the G25 datasheets at the usual link here.
A basic distance analysis with the G25 data at Vahaduo shows that the two samples labeled Himera_480BCE_3 are either early Balts or Slavs. I suspect that they're Slavs, because I believe that early Slavs had this type of Baltic-like genetic structure before mixing with their non-Slavic-speaking neighbors. Well, that's my pet theory for now, so take it or leave it.
Distance to: ITA_Sicily_Himera_480BCE_3:I10943 0.03393838 HUN_IA_La_Tene_o:I18226 0.03572886 DEU_MA_Krakauer_Berg:KRA001 0.03618075 RUS_Pskov_VA:VK159 0.03899963 SWE_Gotland_VA:VK463 0.03915018 Baltic_EST_IA:s19_V12_1 Distance to: ITA_Sicily_Himera_480BCE_3:I10949 0.03573636 HUN_IA_La_Tene_o3:I25524 0.03698768 HUN_IA_La_Tene_o:I18226 0.03732752 SWE_Skara_VA:VK397 0.03767022 Baltic_EST_IA:s19_V12_1 0.03772687 DEU_MA_Krakauer_Berg:KRA001On the other hand, I'm almost certain that the two Himera_480BCE_4 samples are Sarmatians. The good old G25 does it again!
Distance to: ITA_Sicily_Himera_480BCE_4:I10944 0.03100861 KAZ_Segizsay_Sarmatian:SGZ002 0.03548059 MDA_Sarmatian:I11925 0.03619219 RUS_Urals_Sarmatian:MJ56 0.03626538 RUS_Urals_Sarmatian:chy001 0.03904260 RUS_Urals_Sarmatian:MJ41 Distance to: ITA_Sicily_Himera_480BCE_4:I10947 0.02989458 RUS_Urals_Sarmatian:MJ43 0.03052790 RUS_Urals_Sarmatian:chy002 0.03170622 KAZ_Kangju:DA226 0.03288789 TUR_BlackSea_Samsun_Anc_C:I4529 0.03310149 KAZ_Aigyrly_Sarmatian:AIG003See also... Slavic-like Medieval Germans
Labels:
ancient ancestry,
ancient DNA,
Ancient Greece,
Balto-Slavic,
battle,
East Baltic,
Eastern Europe,
Eastern European steppe,
Himera,
Italy,
Mediterranean,
Northeastern Europe,
Sarmatian,
Sicily,
Slavic,
Slavs
Monday, September 19, 2022
Dear Iosif...Yamnaya
Even though the Yamnaya culture probably originated in what is now Ukraine, the earliest Yamnaya samples currently available are from the modern-day Samara region of Russia. They mostly date to around 3,000 BCE. I can analyze their ancestry using Principal Component Analysis (PCA) data.
Target: RUS_Yamnaya_Samara Distance: 3.2816% / 0.03281581 81.0 RUS_Progress_En 14.4 UKR_N 4.6 HUN_Vinca_MN 0.0 ARM_Aknashen_N 0.0 ARM_Masis_Blur_N 0.0 AZE_Caucasus_lowlands_LN 0.0 BGR_C 0.0 BGR_Dzhulyunitsa_N 0.0 IRN_Ganj_Dareh_N 0.0 IRN_Hajji_Firuz_C 0.0 IRN_Seh_Gabi_C 0.0 IRN_Tepe_Abdul_Hosein_N 0.0 IRN_Wezmeh_N 0.0 RUS_Darkveti-Meshoko_En 0.0 RUS_Maykop 0.0 RUS_Maykop_Late 0.0 RUS_Maykop_NovosvobodnayaThe above results show exactly zero ancestry from West Asia. Admittedly, both RUS_Progress_En and HUN_Vinca_MN are European ancients with significant West Asian-related ancestry. However, this ancestry is very distantly West Asian-related, and, for instance, it almost certainly has no relevance to the Indo-Anatolian homeland debate. The Afanasievo culture of Central Asia is regarded to have been an early offshoot of the Yamnaya culture. A good number of Afanasievo samples are available, so let's have a look if their results match those of the Yamnaya folks. And indeed they do, since BGR_C is very similar to HUN_Vinca_MN.
Target: RUS_Afanasievo Distance: 3.4055% / 0.03405499 84.0 RUS_Progress_En 11.4 UKR_N 4.6 BGR_C 0.0 ARM_Aknashen_N 0.0 ARM_Masis_Blur_N 0.0 AZE_Caucasus_lowlands_LN 0.0 BGR_Dzhulyunitsa_N 0.0 HUN_Vinca_MN 0.0 IRN_Ganj_Dareh_N 0.0 IRN_Hajji_Firuz_C 0.0 IRN_Seh_Gabi_C 0.0 IRN_Tepe_Abdul_Hosein_N 0.0 IRN_Wezmeh_N 0.0 RUS_Darkveti-Meshoko_En 0.0 RUS_Maykop 0.0 RUS_Maykop_Late 0.0 RUS_Maykop_NovosvobodnayaTo try this at home, stick the PCA data in the text file here into the relevant fields here and cranck up the "Cycles" to 4X. You should see exactly zero ancestry from West Asia every time. I can, more or less, reproduce these results with tools that are routinely used in peer reviewed papers. Below is a table of mixture models produced with the qpAdm software. I set the pass threshold to P ≥0.05, which is an arbitrary value, but the pattern is clear. The full output from each qpAdm run is available here. Importantly, qpAdm needs to be fed the relevant "right pop" outgroups to be able to discriminate accurately between reference populations.
right pops: CMR_Shum_Laka_8000BP MAR_Taforalt Levant_Natufian IRN_Ganj_Dareh_N Levant_PPNB TUR_Marmara_Barcin_N HUN_Starcevo_N HUN_Koros_N SRB_Iron_Gates_HG Iberia_Southeast_Meso RUS_Karelia_HG RUS_West_Siberia_HG RUS_Boisman_MN MNG_North_N TWN_Hanben BRA_LapaDoSanto_9600BPSo, for instance, if one were to use in this role the modern-day Mbuti people, as opposed to, say, the ancient hunter-gatherers of Shum Laka, one might find that many models look statistically better than they should. And then one might also find that the Yamnaya samples carry significant West Asian ancestry. Actually, I'm not opposed to the idea of some West Asian ancestry in Yamnaya. Indeed, considering the extraordinary mobility of the Yamnaya people and their Eneolithic predecessors on the Pontic-Caspian steppe, it would be unusual if they didn't come into close contact and mix, to some degree, with their neighbors from West Asia. However, based on everything I've seen, from uniparental markers to different types of autosomal genetic tests, it's clear to me that there's no substantial West Asian ancestry in any Yamnaya samples, except for an outlier female from modern-day Ozera, Ukraine (see here). Admittedly, ancient DNA does have a habit of throwing curveballs, so I'm eagerly awaiting new Eneolithic samples from the Pontic-Caspian steppe, particularly those associated with the Yamnaya-like Sredni Stog culture, to help finally settle this issue. Believe it or not, a contact recently sent me a supposedly unpublished female sample from a ~4,200 BCE Sredni Stog burial in modern-day Igren, east central Ukraine. So what the hell, let's assume for the time being that this sample is genuine. This is how Miss Sredni Stog behaves in my PCA mixture test.
Target: UKR_Sredni_Stog Distance: 4.0769% / 0.04076877 75.6 RUS_Progress_En 17.8 UKR_N 6.6 HUN_Vinca_MN 0.0 ARM_Aknashen_N 0.0 ARM_Masis_Blur_N 0.0 AZE_Caucasus_lowlands_LN 0.0 BGR_C 0.0 BGR_Dzhulyunitsa_N 0.0 HUN_Vinca_MN 0.0 IRN_Ganj_Dareh_N 0.0 IRN_Hajji_Firuz_C 0.0 IRN_Seh_Gabi_C 0.0 IRN_Tepe_Abdul_Hosein_N 0.0 IRN_Wezmeh_N 0.0 RUS_Darkveti-Meshoko_En 0.0 RUS_Maykop 0.0 RUS_Maykop_Late 0.0 RUS_Maykop_NovosvobodnayaWow, just wow. Have we actually found Miss Proto-Yamnaya? What does qpAdm have to say in the matter?
UKR_Sredni_Stog HUN_Vinca_MN 0.034±0.028 RUS_Progress_En 0.796±0.045 UKR_N 0.170±0.034 P-value 0.41088Again, this is an excellent match with the results from my PCA test, especially if we take into account the standard errors. However, with qpAdm it's also possible to model this individual's ancestry as part West Asian.
UKR_Sredni_Stog AZE_Caucasus_lowlands_LN 0.056±0.039 RUS_Progress_En 0.761±0.061 UKR_N 0.183±0.036 P-value 0.465667As I pointed out above, it's plausible for such people to harbor some West Asian ancestry, but I'm very sceptical that this is really the case here, despite the rather solid qpAdm statistical fit. That's because UKR_Sredni_Stog is not a high quality sample, and, from my experience, qpAdm often has problems analyzing fine scale ancestry in singletons or even small groups that show excess DNA damage and/or offer much less than a million markers. See also... Dear Iosif, about that ~2% But Iosif, what about the Phrygians?
Friday, September 9, 2022
Dear Iosif, about that ~2%
The debate over the location of the so called Indo-Anatolian homeland won't be decided by the persistence of any type of genetic ancestry in ancient Anatolia.
It'll be decided by a multidisciplinary study on the interactions between the ancient peoples of the North Pontic steppe, the eastern Balkans, and western Anatolia.
If such a study finds a pulse of steppe-related gene flow from the Balkans into Anatolia sometime during the early metal ages, it'll corroborate the linguistic hypothesis that a language ancestral to Hittite, Luwian and related tongues moved into Anatolia from Eastern Europe.
Why do we only need a pulse of gene flow, you might ask? Obviously, because:
- language and genetic ancestry can start with a strong association but, since they're not linked, they can eventually follow very different trajectories - the dilution of genetic ancestry is an important factor, especially in ancient West Asia, and it must be taken into account in models of language spread, rather than ignored in favor of simple, elegant models that do not reflect reality.Here's my favorite quote from the recent Lazaridis, Alpaslan-Roodenberg et al. paper, because, probably unbeknownst to the authors, it's exceptionally revealing about the spread of a wide range of Indo-European speakers into Anatolia.
However, in individuals from Gordion, a Central Anatolian city that was under the control of Hittites before becoming the Phrygian capital and then coming under the control of Persian and Hellenistic rulers, the proportion of Eastern hunter-gatherer ancestry is only ~2%, a tiny fraction for a region controlled by at least four different Indo-European–speaking groups.Indeed, this is exactly what the Lazaridis, Alpaslan-Roodenberg et al. paper should've been about. That is, the authors should've given us a painstaking account of the spread of different ancient Indo-European speaking groups into Anatolia and explained how, overall, their DNA was rapidly diluted to a trace amount. However, instead they treated us to a make-believe tale about a so called Indo-Anatolian homeland in what is now Armenia. See also... Dear Iosif...Yamnaya But Iosif, what about the Phrygians? Dear Iosif... Dear Iosif #2 Dear Iosif #3
Sunday, September 4, 2022
But Iosif, what about the Phrygians?
A paper in Science co-authored by around 200 scientists from some of the world's top academic institutions surely must mean something, right? Not necessarily.
In this short blog post I'll try to explain, as simply as I can, why the Lazaridis, Alpaslan-Roodenberg et al. paper doesn't get us any closer to solving the riddle of the so called Indo-Anatolian homeland.
However, it must be said that the paper does include many interesting and valuable samples. I'll be using six of these samples, labeled TUR_C_Gordion_Anc, to argue my case.
The TUR_C_Gordion_Anc sample set is from Gordion, the capital of ancient Phrygia, and thus, in all likeliness, it represents Phrygian speakers.
Phrygian is an Indo-European language and the leading hypothesis is that it originated in the Balkans.
In terms of fine scale ancestry, TUR_C_Gordion_Anc can be reliably divided into two genetic clusters. In the Principal Component Analysis (PCA) below these clusters are labeled TUR_C_Gordion_Anc1 and TUR_C_Gordion_Anc2.
Note that TUR_C_Gordion_Anc1 is obviously pulling away from TUR_C_Gordion_Anc2 towards samples from the Balkans. I've used ancient samples from what is now North Macedonia, labeled MKD_Anc, to represent the Balkans. To see an interactive version of the plot, paste the PCA coordinates from here into the relevant field here.
Visually, this is not an especially dramatic outcome, but it's an incredible result nonetheless, because it shows that even a few ancient samples can help to solve an age old mystery.
Across many dimensions of genetic variation, the shift in the PCA from TUR_C_Gordion_Anc1 to TUR_C_Gordion_Anc2 represents about 20% admixture from the Balkans, and about 8% from the Eastern European steppe. That's plenty enough to corroborate the linguistic hypothesis that the Phrygians originated in the Balkans, and that some of their ancestors came from the steppe. The mixture models below were done with the tools here.
Target: TUR_C_Gordion_Anc1 Distance: 1.6634% / 0.01663373 40.6 Kura-Araxes_ARM_Kaps 22.2 Anatolia_Barcin_N 21.8 MKD_Anc 13.6 Levant_PPNB 1.4 IRN_Ganj_Dareh_N 0.4 Han Target: TUR_C_Gordion_Anc1 Distance: 1.7109% / 0.01710904 40.2 Kura-Araxes_ARM_Kaps 37.8 Anatolia_Barcin_N 12.4 Levant_PPNB 8.0 Yamnaya_RUS_Samara 1.2 IRN_Ganj_Dareh_N 0.4 Han Target: TUR_C_Gordion_Anc2 Distance: 2.0293% / 0.02029339 51.0 Kura-Araxes_ARM_Kaps 26.8 Anatolia_Tepecik_Ciftlik_N 17.6 Anatolia_Barcin_N 4.6 Levant_PPNBSurprisingly, Lazaridis, Alpaslan-Roodenberg et al. didn't have much to say about this topic. This quote basically sums it up:
However, in individuals from Gordion, a Central Anatolian city that was under the control of Hittites before becoming the Phrygian capital and then coming under the control of Persian and Hellenistic rulers, the proportion of Eastern hunter-gatherer ancestry is only ~2%, a tiny fraction for a region controlled by at least four different Indo-European–speaking groups.I have no doubt that Lazaridis, Alpaslan-Roodenberg et al. can run a very decent PCA, and then blow it up to a size big enough to show that the Gordion samples represent two genetically somewhat distinct groups. I'm also sure that, if they really try, they can locate significant levels of proximate and relevant European ancestry in some of these samples. They don't have to use my methods; they can use any methods they like. My point is that they won't find much if they're just looking for genetic signals from the Upper Paleolithic or Mesolithic. Now, considering the way that the Phrygian question was treated by Lazaridis, Alpaslan-Roodenberg et al., despite the fact that they managed to sequence a few likely Phrygian speakers from none other than the Phrygian capital, let's not pretend that their paper brought us any closer to understanding the genetic origins of Anatolian speakers or pinpointing their ancestral homeland. In order to even try to solve these problems with ancient DNA, we need a wide range of samples from Hittite, Luwian and other key sites where Anatolian languages were spoken. And then we must analyze them properly. I'm guessing that Lazaridis, Alpaslan-Roodenberg et al. went out of their way to get such samples, but for one reason or another they failed. If so, that's OK, but I have a feeling that even if they got them, they wouldn't know what to do with them, because at best these samples would only show ~2% Eastern hunter-gatherer ancestry. Haha. For what it's worth, I believe that the ancient data in the Lazaridis, Alpaslan-Roodenberg et al. paper point to the North Pontic steppe as the Indo-Anatolian homeland, and I'll lay out my arguments in an upcoming blog post. See also... Dear Iosif...Yamnaya Dear Iosif, about that ~2% Dear Iosif... Dear Iosif #2 Dear Iosif #3
Thursday, September 1, 2022
Dear Iosif #3
Back in 2016 I made this prediction about the origins of the Yamnaya people (Steppe_EMBA):
But here's my prediction: Steppe_EMBA only has 10-15% admixture from the post-Mesolithic Near East not including the North Caucasus, and basically all of this comes via female mediated gene flow from farming communities in the Caucasus and perhaps present-day Ukraine.The relevant blog post is still here. Looking back, my analysis is a bit sloppy and I didn't articulate my ideas too well. But that was a pretty good prediction for its time, and I believe it still has a chance of being confirmed, more or less. On the other hand, the widely publicized hypothesis that the Yamnaya population is a ~50/50 mixture between indigenous Eastern European hunter-gatherers and Near Eastern or West Asian migrants never looked right to me. So I'm glad that it's now dead and buried. Those of you not up to date with this topic, all you need to know is that the Yamnaya genotype existed in Eastern Europe at least a thousand years before Yamnaya, and, moreover, the Yamnaya people are largely derived from Eastern European foragers already rich in Near Eastern-related ancestry. The relevant ancient genomes are on the way (for instance, see here). Nevertheless, the narrative that waves of Near Eastern migrants moved into prehistoric Eastern Europe, leading to the emergence of the Yamnaya culture and even the Proto-Indo-European language, is still being pushed by some notable scientists working with ancient DNA. My hope is that, considering the latest revelations about the genetic origins of the Yamnaya people, these scientists can embrace a more nuanced view. How about something like this?
- people moved around, and they were especially mobile on the Eastern European steppe from the Eneolithic onwards - when they made contact they sometimes mixed, so there was admixture between far flung steppe groups - since population densities on the steppe were low until the Yamnaya period, minor admixture that entered the steppe during the Neolithic and Eneolithic wasn't dilluted easily.See also... Dear Iosif #2 But Iosif, what about the Phrygians? Dear Iosif, about that ~2% Dear Iosif...Yamnaya
Sunday, August 28, 2022
Dear Iosif #2
In my last blog post I made a mistake in my interpretation of this quote from Lazaridis, Alpaslan-Roodenberg et al., because it confused the crap out of me:
However, the complete lack of association of R-haplogroup descendants and EHG ancestry in either Armenia or Iran is consistent with either a massive dilution of EHG ancestry in these populations resulting in the dissociation of Y-chromosome lineages from autosomal ancestry over time, or with a scenario in which R-M269 was not associated with substantial EHG ancestry to begin with.I thought they meant that they couldn't find any Eastern European hunter-gatherer (EHG) ancestry in samples from Armenia or Iran bearing Y-chromosome R1b-M269. Of course, they did find EHG ancestry in these individuals, it's just that they couldn't establish an association specifically between this type of ancestry and Y-haplogroup R1b. That is, males with Y-haplogroup R1b in Armenia, Iran and everywhere else generally show about the same level of EHG ancestry as their ethnic kin with other Y-haplogroups. But so what? Why mention this when discussing the origins of R1b-M269, when it has absolutely no value in this context? Y-haplogroups aren't linked directly to autosomal DNA, and Lazaridis, Alpaslan-Roodenberg et al. are obviously aware of this (hence their point about the potential massive dilution of EHG ancestry). In regards to the origins of R1b-M269, and the provenance of West Asian R1b-M269, the really powerful observation is that R1b-M269 shows up rather late and suddenly in the West Asian ancient DNA record along with EHG and steppe ancestry. That, and the fact that Eastern Europe is an ancient R1b hotbed (while West Asia a desert), means there's virtually no chance that R1b-M269 is native to West Asia. In other words, there was no R1b-M269 in West Asia until the steppe people brought it there from north of the Caucasus. See also... Dear Iosif... Dear Iosif #3 But Iosif, what about the Phrygians? Dear Iosif, about that ~2% Dear Iosif...Yamnaya
Saturday, August 27, 2022
Dear Iosif...
Update 29/08/22: Dear Iosif #2
...
I'm skimming through the Lazaridis, Alpaslan-Roodenberg et al. paper that just came out at Science. And I feel like someone punched me in the face.
Nevertheless, I'll try to be diplomatic. Suffice to say, for now, that there's some rather strange stuff in this paper.
The main problem is that the authors are attempting to study fine scale ancestry with a somewhat rough distal model. As a result, they miss important details.
For instance, this quote is from the paper's supplementary PDF file, freely available here.
However, the complete lack of association of R-haplogroup descendants and EHG ancestry in either Armenia or Iran is consistent with either a massive dilution of EHG ancestry in these populations resulting in the dissociation of Y-chromosome lineages from autosomal ancestry over time, or with a scenario in which R-M269 was not associated with substantial EHG ancestry to begin with.Obviously, EHG means Eastern European Hunter-Gatherer. But why focus on EHG? Surely, this makes little sense when looking at the genetic prehistory of West Asia, because no one ever argued that this region was settled by EHG populations. It was widely settled by Yamnaya-related groups, with already heavily diluted EHG ancestry, during the metal ages. OK, so the authors are actually aware about the potential dilution of EHG ancestry, but they don't really do anything about it. If we're looking at the origins of West Asian R1b-M269, and using its association with autosomal DNA components as a guide, then we should be focusing on Yamnaya-related ancestry. For instance, here's a fine scale ancient ancestry model based on Principal Component Analysis (PCA) data. It shows the ancestry proportions of two relatively high coverage Iron Age males from two different sites in Iran from the Lazaridis, Alpaslan-Roodenberg et al. dataset. Both belong to R1b-M269 and both show significant Yamnaya-related ancestry.
Target: IRN_HajjiFiruz_IA:I2327_all Distance: 2.2930% / 0.02292994 39.6 Kura-Araxes_ARM_Kaps 24.2 IRN_Ganj_Dareh_N 18.2 Levant_PPNB 12.4 Yamnaya_RUS_Samara 4.4 Anatolia_Tepecik_Ciftlik_N 1.2 Han Target: IRN_Hasanlu_IA:I4232_all Distance: 2.5179% / 0.02517895 26.0 IRN_Ganj_Dareh_N 25.6 Kura-Araxes_ARM_Kaps 24.4 Anatolia_Tepecik_Ciftlik_N 15.8 Yamnaya_RUS_Samara 7.6 Levant_PPNB 0.6 IRN_Shahr_I_Sokhta_BA2As a control, here's an earlier, Chalcolithic sample bearing Y-haplogroup J2b from the same region. Not surprisingly, this individual totally lacks the Yamnaya-related signal.
Target: IRN_HajjiFiruz_ChL:I4241_all Distance: 2.7938% / 0.02793782 32.6 Kura-Araxes_ARM_Kaps 25.6 IRN_Ganj_Dareh_N 23.6 Anatolia_Tepecik_Ciftlik_N 18.2 Levant_PPNBOverall, these results make perfect sense. I could probably locate very minor signals of EHG ancestry in the Iron Age samples, but that would be more difficult and much less certain, so I won't bother. Soon I'll be able to rerun these analyses with Bronze Age samples from Dagestan and surrounds. That should bump up the levels of Yamnaya-related ancestry and improve the statistical fits (wink, wink, nudge, nudge). Disappointingly, Lazaridis, Alpaslan-Roodenberg et al. go so far as to suggest that R1b-M269 may have originated in West Asia. However, considering the scores of ancient Eastern European populations rich in R1b-M269 and many near and far related subclades of R1b, this makes no sense whatsoever. Indeed, contemplating nowadays that R1b-M269 might be native to West Asia, where R1b only starts showing up in the ancient DNA record during the Copper Age, is about as stupid as claiming that gravity doesn't exist. Largely due to their distal model approach, Lazaridis, Alpaslan-Roodenberg et al. also argue that the Indo-Anatolian homeland was located in what is now Armenia and surrounds. I'm far from convinced that this solution will stand the test of time. In terms of the more widely accepted theory that the Indo-Anatolian homeland was located on the Pontic-Caspian steppe in Eastern Europe, the most important samples in the paper are the three Bronze Age individuals from Yassitepe in western Anatolia. That's because they're from a region that is traditionally seen as the entry point of Indo-Anatolian speakers into Anatolia from the European steppe via the Balkans. Interestingly, individual I5737, dated to 2035-1900 calBCE or the Middle Bronze Age, belongs to Y-chromosome haplogroup I2a-P78, which surely must be a signal of European ancestry. I see this as a significant result. Here's how the trio from Yassitepe look in my fine scale ancient ancestry model. Minor Yamnaya-related ancestry does show up, although, admittedly, it might just be noise in individual I5735.
Target: TUR_Aegean_Izmir_Yassitepe_MBA:I5737 Distance: 2.7507% / 0.02750748 58.4 Anatolia_Barcin_N 20.4 Kura-Araxes_ARM_Kaps 9.2 Anatolia_Tepecik_Ciftlik_N 5.6 IRN_Ganj_Dareh_N 3.8 Yamnaya_RUS_Samara 2.6 Levant_PPNB Target: TUR_Aegean_Izmir_Yassıtepe_EBA:I5733 Distance: 2.7969% / 0.02796887 52.0 Anatolia_Barcin_N 27.2 Kura-Araxes_ARM_Kaps 8.6 Levant_PPNB 6.2 Yamnaya_RUS_Samara 6.0 IRN_Ganj_Dareh_N Target: TUR_Aegean_Izmir_Yassıtepe_EBA:I5735 Distance: 3.1270% / 0.03127009 36.0 Kura-Araxes_ARM_Kaps 32.4 Anatolia_Tepecik_Ciftlik_N 26.0 Anatolia_Barcin_N 2.8 IRN_Shahr_I_Sokhta_BA2 1.2 Yamnaya_RUS_Samara 1.0 Levant_PPNB 0.6 MAR_TaforaltThis isn't much, especially considering it's already late 2022, but it's better than nothing. Fortunately, more samples from Bronze Age western Anatolia are on the way (wink, wink, nudge, nudge). However, I'm not done with the Lazaridis, Alpaslan-Roodenberg et al. dataset yet. I'm planning to spend much more time on this blog in the coming weeks and months and will be using their samples in a wide range of analyses. Citation... Iosif Lazaridis, Songül Alpaslan-Roodenberg et al., The genetic history of the Southern Arc:A bridge between West Asia and Europe, Science 377, eabm4247 (2022) See also... Dear Iosif #3 But Iosif, what about the Phrygians? Dear Iosif, about that ~2% Dear Iosif...Yamnaya
Labels:
Anatolia,
ancient ancestry,
ancient DNA,
Caucasus,
Hittite,
Indo-Anatolian,
Indo-European,
Iosif Lazaridis,
Iran,
Luwian,
Pontic-Caspian steppe,
Proto-Indo-European,
R1b-M269,
Turkey,
Ukraine,
Yamnaya
Friday, August 12, 2022
Mediterranean PCA update
I updated my Principal Component Analysis (PCA) of Mediterranean populations with most of the ancient Jewish samples from the Waldman et al. preprint. To view intereactive versions of the plots paste the data from here into the PCA DATA field here and press PLOT PCA. The ancient Jews are labeled DEU_MA_Erfurt.
See anything interesting? I'm again seeing more complexity than claimed by Waldman et al., but what would I know anyway?
See also...
My take on the Erfurt Jews
Greeks in a Longobard cemetery
Tuesday, June 21, 2022
My take on the Erfurt Jews
I had a quick look at the genotype data from the recent Waldman et al. preprint focusing on the ancestry of early Jews from Erfurt, Germany. My impression is that the genetic origins of these Jews are somewhat more complex than claimed in the manuscript.
Indeed, I'd say the Waldman et al. characterization of the Erfurt Jews as a three-way mixture between populations similar to present-day Lebanese, South Italians and Russians doesn't exactly reflect reality.
Unlike Waldman et al., I designed an ADMIXTURE analysis that separated East Asian ancestry into East Asian and Siberian clusters, and also included Mediterranean and North African clusters. The output is available in a spreadsheet HERE. Below is a bar graph based on some of the output.
Now, keeping in mind that ADMIXTURE is not a formal mixture test, and that it estimates ancestry proportions from inferred populations, as opposed to ancient groups that actually existed, here are some key observations:
- in terms of fine scale ancestry, the Erfurt Jews show enough variation to be divided into three or four clusters, as opposed to just two as per Waldman et al. - some of the Erfurt Jews show excess "Mediterranean" ancestry, while others excess "North African" ancestry, and this cannot be explained with ancestral populations similar to Lebanese and/or South Italians, but rather with significant gene flow from the western Mediterranean and possibly North Africa - several of the Erfurt Jews show relatively high levels of "East Asian" ancestry that cannot be explained by admixture from Russians, or even any Russian-like populations, because such populations almost lack this type of ancestry, and instead show significant "Siberian" admixture - as far as I can see, there are no correlations between any of the observations above and the quality of the samples. That is, low coverage doesn't appear to be causing the aforementioned excess "Mediterranean", "North African" and/or "East Asian" ancestry proportions.Investigating this in more detail with, say, formal statistics will take some time. But I was able to reproduce the results from the above ADMIXTURE run using several somewhat different datasets, so that's something. It seems to me that Waldman et al. want a simple and elegant model to explain the data, which is understandable, but I do think they should at least expand their ADMIXTURE analysis to include "Siberian", "Mediterranean" and "North African" clusters, and go from there depending on what they find. Citation... Waldman et al., Genome-wide data from medieval German Jews show that the Ashkenazi founder event pre-dated the 14th century, bioRxiv, posted May 16, 2022, doi: https://doi.org/10.1101/2022.05.13.491805 See also... Mediterranean PCA update
Labels:
admixture,
ancient ancestry,
ancient DNA,
Ashkenazi,
Central Europe,
East Asia,
Eastern Europe,
Erfurt Jews,
Germany,
Jewish,
Jews,
Medieval,
Mediterranean,
Middle Ages,
North Africa,
Poland,
Russia,
Slavic,
Slavs
Saturday, June 18, 2022
David Reich on the origin of the Yamnaya people (!?)
Harvard's David Reich is doing a talk next month about the genetic history of West Asia and nearby parts of Europe. This is a quote from an online abstract of the talk (found here).
The impermeability of Anatolia to exogenous migration contrasts with our finding that the Yamnaya had two distinct gene flows, both from West Asia, suggesting that the Indo-Anatolian language family originated in the eastern wing of the Southern Arc and that the steppe served only as a secondary staging area of Indo-European language dispersal.If this is actually what David Reich is going to claim then I'd say his team has a lot of work to do before they put out their paper on the topic. First of all, Yamnaya did not have two distinct gene flows from West Asia. I don't even know what that means exactly, but there's no way that this statement is correct no matter how one interprets it. In fact, the Yamnaya population formed on the Pontic-Caspian steppe from earlier groups native to this part of Eastern Europe, such as the people associated with the Sredny Stog culture. That is, there were no migrations from West Asia into Eastern Europe that can be claimed to have been instrumental in the emergence of the Yamnaya population. On the other hand, Yamnaya may have been significantly influenced by cultural impulses from West Asia, but this is nothing new. In terms of deep population structure, the Yamnaya genotype can be described as a mixture between Eastern European and West Asian-related genetic components. However, these Asian-related components were already in Europe thousands of years before Yamnaya came into existence. Indeed, soon to be published ancient DNA shows that hunter-gatherers very similar to the Yamnaya people, packing quite a lot of West Asian-related ancestry, lived in the Middle Don region (just north of the Pontic-Caspian steppe) well before 5,000 BCE (see here). So, did the West Asian ancestors of these Middle Don hunter-gatherers speak Proto-Indo-European, or, as David Reich calls it, Indo-Anatolian? Keep in mind that most linguists put the birth of Indo-Anatolian around 4,000 BCE, which is actually the Sredny Stog period. Moreover, in underlining Anatolia's supposed impermeability to exogenous migration, David Reich is arguing against things that no one worth their salt ever claimed. That's because the spread of Indo-Anatolian speakers into Anatolia has never really been described by archeologists and linguists as a massive migration, but rather as an infiltration into lands already heavily populated by the Hattians (for instance, see here). We may have already seen the genetic evidence of this infiltration in the presence of steppe Y-chromosome haplogroup R-V1636 in a Chalcolithic burial at Arslantepe (see here and here). Let's wait and see what else crops up over the next few years as many more ancient Anatolian genomes are sequenced by David Reich and colleagues. See also...
Labels:
Anatolia,
ancient DNA,
David Reich,
David Reich Lab,
Eastern Europe,
Harvard,
Hittite,
Indo-Anatolian,
PIE,
PIE homeland,
Pontic-Caspian steppe,
Proto-Indo-European,
West Asia,
Yamnaya
Wednesday, May 18, 2022
Geography is hard (for some)
It's that time of the academic year again when bioRxiv is inundated with ancient DNA preprints. I'm not complaining, but I almost spat out my coffee when I saw this map in one of the new manuscripts (here).
What's the logic behind labeling almost all of Eastern Europe as "Steppe", and instead labeling just Czechia, Hungary and Slovakia as "Eastern Europe"? In my opinion those three countries, plus Poland, are better described as East Central Europe anyway.
It seems to me that many people working at the highest level in population genetics simply don't know what the Eurasian steppe is. They appear to see it as a continent of its own, when, in fact, it's a topographical feature and ecoregion that straddles the continents of Europe and Asia. That's why it's called the Eurasian steppe, and it's made up of three main parts: the Pontic-Caspian steppe of Eastern Europe, the Kazkah steppe of Central Asia, and the Eastern steppe of Mongolia.
Here's the same map with a few corrections (in red). Much better, don't you think?
Citation...
Antonio et al., Stable population structure in Europe since the Iron Age, despite high mobility, bioRxiv, posted May 16, 2022, doi: https://doi.org/10.1101/2022.05.15.491973
See also...
Matters of geography
Tuesday, May 17, 2022
Genome-wide data from medieval German Jews (Waldman et al. 2022 preprint)
Over at bioRxiv at this LINK. Here's the abstract:
We report genome-wide data for 33 Ashkenazi Jews (AJ), dated to the 14th century, following a salvage excavation at the medieval Jewish cemetery of Erfurt, Germany. The Erfurt individuals are genetically similar to modern AJ and have substantial Southern European ancestry, but they show more variability in Eastern European-related ancestry than modern AJ. A third of the Erfurt individuals carried the same nearly-AJ-specific mitochondrial haplogroup and eight carried pathogenic variants known to affect AJ today. These observations, together with high levels of runs of homozygosity, suggest that the Erfurt community had already experienced the major reduction in size that affected modern AJ. However, the Erfurt bottleneck was more severe, implying substructure in medieval AJ. Together, our results suggest that the AJ founder event and the acquisition of the main sources of ancestry pre-dated the 14th century and highlight late medieval genetic heterogeneity no longer present in modern AJ.It's nice to finally see some ancient Jewish genotypes on the way, but there's a bit of a problem with this preprint. The fact that the authors are using modern-day Russians to model Eastern European-related ancestry in these Ashkenazi ancients from Central Europe tells me that they're somewhat confused. They did this because some of the Jews harbor significant Slavic ancestry and minor but perceptible East Asian ancestry, and Russians are Slavs who carry some Siberian ancestry, which is closely related to East Asian ancestry. Thus, broadly speaking, in terms of the right mix of DNA, Russians do the job. However, as per the preprint, based on historical data, these Jews probably sourced their Slavic ancestry from Bohemia, Moravia and/or Silesia, and the Slavic speakers in these regions carry very little, if any, East Asian or Siberian ancestry. I'm sure the authors can verify this claim without too much trouble. Ergo, it's likely that the Erfurt Jews received their Slavic and East Asian admixtures from different sources, and possibly at different times. I'd like to see Waldman et al. tackle this issue properly. I suspect that if they do, they might discover something interesting and perhaps unexpected about the ethnogenesis of Ashkenazi Jews. See also... My take on the Erfurt Jews Mediterranean PCA update
Saturday, May 7, 2022
Population genomics of Stone Age Eurasia (Allentoft et al. 2022 preprint)
Over at bioRxiv at this LINK. It'll take me a few days to read this manuscript properly. Here's the abstract:
The transitions from foraging to farming and later to pastoralism in Stone Age Eurasia (c. 11-3 thousand years before present, BP) represent some of the most dramatic lifestyle changes in human evolution. We sequenced 317 genomes of primarily Mesolithic and Neolithic individuals from across Eurasia combined with radiocarbon dates, stable isotope data, and pollen records. Genome imputation and co-analysis with previously published shotgun sequencing data resulted in >1600 complete ancient genome sequences offering fine-grained resolution into the Stone Age populations. We observe that: 1) Hunter-gatherer groups were more genetically diverse than previously known, and deeply divergent between western and eastern Eurasia. 2) We identify hitherto genetically undescribed hunter-gatherers from the Middle Don region that contributed ancestry to the later Yamnaya steppe pastoralists; 3) The genetic impact of the Neolithic transition was highly distinct, east and west of a boundary zone extending from the Black Sea to the Baltic. Large-scale shifts in genetic ancestry occurred to the west of this "Great Divide", including an almost complete replacement of hunter-gatherers in Denmark, while no substantial ancestry shifts took place during the same period to the east. This difference is also reflected in genetic relatedness within the populations, decreasing substantially in the west but not in the east where it remained high until c. 4,000 BP; 4) The second major genetic transformation around 5,000 BP happened at a much faster pace with Steppe-related ancestry reaching most parts of Europe within 1,000-years. Local Neolithic farmers admixed with incoming pastoralists in eastern, western, and southern Europe whereas Scandinavia experienced another near-complete population replacement. Similar dramatic turnover-patterns are evident in western Siberia; 5) Extensive regional differences in the ancestry components involved in these early events remain visible to this day, even within countries. Neolithic farmer ancestry is highest in southern and eastern England while Steppe-related ancestry is highest in the Celtic populations of Scotland, Wales, and Cornwall (this research has been conducted using the UK Biobank resource); 6) Shifts in diet, lifestyle and environment introduced new selection pressures involving at least 21 genomic regions. Most such variants were not universally selected across populations but were only advantageous in particular ancestral backgrounds. Contrary to previous claims, we find that selection on the FADS regions, associated with fatty acid metabolism, began before the Neolithisation of Europe. Similarly, the lactase persistence allele started increasing in frequency before the expansion of Steppe-related groups into Europe and has continued to increase up to the present. Along the genetic cline separating Mesolithic hunter-gatherers from Neolithic farmers, we find significant correlations with trait associations related to skin disorders, diet and lifestyle and mental health status, suggesting marked phenotypic differences between these groups with very different lifestyles. This work provides new insights into major transformations in recent human evolution, elucidating the complex interplay between selection and admixture that shaped patterns of genetic variation in modern populations.Allentoft et al., Population Genomics of Stone Age Eurasia, bioRxiv, posted May 06, 2022, doi: https://doi.org/10.1101/2022.05.04.490594 See also... Understanding the Eneolithic steppe
Saturday, March 12, 2022
Lousy intel
I don't like discussing current events and politics here, but it's impossible to ignore what is happening in Eastern Europe.
It's a tragedy and catastrophe for both Ukraine and Russia. It's also likely to have a negative impact on ancient DNA research, Indo-European studies, and thus also on this blog.
I'm seeing a lot of confusion online about why Russia invaded Ukraine, but I don't think it's very complicated.
After getting the better of the West in recent years, Russia finally overreached and made a massive tactical blunder, in large part because of lousy intel. More broadly, I also see this as the Soviet Union's dead cat bounce moment.
Russia will now have to reinvent itself, possibly as China's junior partner or even vassal state.
As for the "special military operation", Russia's initial plan was to achieve a quick, relatively bloodless victory, followed by a military parade in Kyiv. But obviously that's not going to happen.
Russia's back up plan, if we can call it that, seems to be to keep pushing into Ukraine at any cost, and hope that the Ukrainians finally tap out. But right now that looks like a long shot.
See also...
Matters of geography
Thursday, February 24, 2022
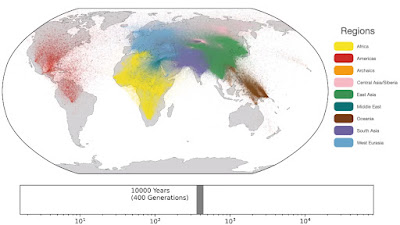
A unified genealogy of modern and ancient genomes (Wilder Wohns et al. 2022)
Over at Science at this LINK. Broadly speaking, this looks like a more sophisticated version of something that I tried about five years ago (see here).
I wonder if they got the idea from me? Honestly, I wouldn't be surprised if they did. But like I say, their methods are way more advanced.
Keep in mind, however, that for now, their analysis includes 3601 modern genomes and just eight ancient genomes. That's because they can only run super high quality ancient sequences. The ratio of ancient genomes will no doubt rise rapidly over the next few years, and that's when things will get really interesting.
Below are some screen caps from a clip accompanying the paper, freely available here. This is the caption to the movie:
Spatio-temporal dynamics in human history. This movie shows the estimated geographic locations of ancestors of Human Genome Diversity Project, Simons Genome Diversity Project, Neanderthal, Denisovan, and Afanasievo samples over time. Each dot represents an edge in the tree sequence of chromosome 20, where the time and geographic location of the parent and child nodes of the edge have been estimated. The locations of edges at each point in time are plotted along the great circle between the parent and child nodes. Edges are colored by the region of the descendants of the child node. If an ancestral lineage has ancestors in multiple regions, its color is the average of the respective colors of each region.See also... Haplotype-based PCA of West Eurasia and Europe
Monday, February 21, 2022
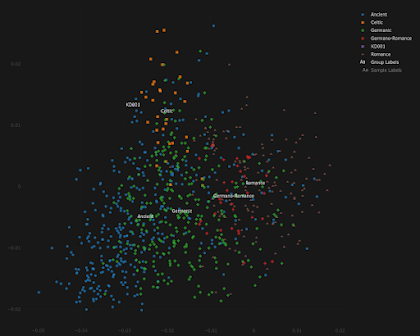
The Pict
KD001 is the first undeniable Pictish sample in my dataset, courtesy of Dulias et al. 2022. Thanks to Altvred for processing the files.
This is how KD001 behaves in my Celtic vs Germanic Principal Component Analysis (PCA). Looks kind of Irish, doesn't he?
To see an interactive version of the plot, paste the coordinates from here into the relevant field here.
See also...
Celtic vs Germanic Europe
Avalon vs Valhalla revisited
When did Celtic languages arrive in Britain?
Labels:
Albion,
ancient DNA,
Britain,
British DNA,
British Isles,
Celtic,
Celtic vs Germanic,
Great Britain,
Indo-European,
Northern Europe,
Northwestern Europe,
Pict,
Pictish,
Picts,
Scotland,
Scottish
Monday, February 14, 2022
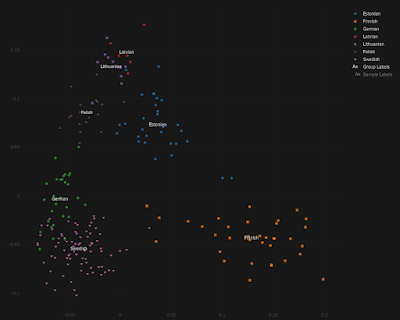
Blond hair is only indirectly associated with Anatolian ancestry in Estonia...duh
In a recent paper about complex traits in Europeans, Marnetto et al. found that blond hair and blue eyes showed a relatively high association with ancient Anatolian ancestry.
This is a somewhat curious finding considering that ancient Anatolians weren't particularly blond haired or blued eyed, and that's probably an understatement.
However, the Europeans that Marnetto et al. based their analysis on were Estonians. And in Estonia ancient Anatolian ancestry peaks in the west and north, probably because this is where Estonians have the most Germanic and Finnish ancestry.
Germanic and Finnish populations are somewhat richer in ancient Anatolian ancestry than Estonians, and, unlike ancient Anatolians, they're often exceptionally blond haired and blue eyed.
So it makes sense that, in Estonia at least, ancient Anatolian ancestry is associated with blond hair and blue eyes, but only indirectly so. The more direct link is between Germanic and Finnish ancestry and blond hair and blue eyes.
I feel that Marnetto et al. should've investigated this, and they also should've made it clear that the associations they found won't necessarily be seen in other European countries.
For the doubters out there, and I know there are at least a few of you, below is a series of Principal Component Analyses (PCA) showing how Estonians compare to other populations from around the Baltic Sea, as well as to present-day Turks from central Anatolia.
Note that, by and large, the same Estonians who show more affinity to the Germanic and/or Finnish individuals are also shifted slightly closer to the Turks, and this is because they harbor elevated ancient Anatolian ancestry. The relevant datasheets are available here.
Citation...
Marnetto et al., Ancestral genomic contributions to complex traits in contemporary Europeans, Current Biology (2022), https://doi.org/10.1016/j.cub.2022.01.046
See also...
Ancient ancestry and complex traits in Estonians (Marnetto et al. 2022)
Mainstream media BS: Europeans owe their height to Asian nomads
Thursday, February 10, 2022
Mainstream media BS: Europeans owe their height to Asian nomads
From a recent Daily Mail article by some clown named Sam Tonkin:
Present day Europeans owe their blue eyes to hunter gatherers, their height to Asian nomads and their blonde hair to Anatolian Neolithic farmers, a new study suggests. ... Most of the contemporary European genetic makeup was shaped by movements that occurred in the last 10,000 years when local hunter gatherers mixed with incoming Anatolian farmers — from present-day Turkey — and Asian nomads, or Pontic Steppe pastoralists. The latter originated from what is now parts of Bulgaria, Romania, Moldova, Ukraine, Russia and Kazakhstan.Haha. Bulgaria, Romania, Moldova and Ukraine are European countries. The relevant parts of Russia and Kazakhstan are also located in Europe. Obviously, the author is referring to the Yamnaya herders who lived on the Pontic-Caspian steppe, which is obviously in Eastern Europe. I blame Johannes Krause for this. See also... Matters of (basic) geography Blond hair is only indirectly associated with Anatolian ancestry in Estonia...duh Ancient ancestry and complex traits in Estonians (Marnetto et al. 2022)
Labels:
ancient DNA,
blond,
blonde hair,
Central Asia,
Daily Mail,
Eastern Europe,
height,
Johannes Krause,
media,
North Pontic steppe,
Pontic-Caspian steppe,
propaganda,
Sam Tonkin,
science,
Yamna,
Yamnaya
Wednesday, February 9, 2022
Ancient ancestry and complex traits in Estonians (Marnetto et al. 2022)
Over at Current Biology at this LINK. Here's the summary:
The contemporary European genetic makeup formed in the last 8,000 years when local Western Hunter-Gatherers (WHGs) mixed with incoming Anatolian Neolithic farmers and Pontic Steppe pastoralists. 1–3 This encounter combined genetic variants with distinct evolutionary histories and, together with new environmental challenges faced by the post-Neolithic Europeans, unlocked novel adaptations. 4 Previous studies inferred phenotypes in these source populations, using either a few single loci 5–7 or polygenic scores based on genome-wide association studies, 8–10 and investigated the strength and timing of natural selection on lactase persistence or height, among others. 6,11,12 However, how ancient populations contributed to present-day phenotypic variation is poorly understood. Here, we investigate how the unique tiling of genetic variants inherited from different ancestral components drives the complex traits landscape of contemporary Europeans and quantify selection patterns associated with these components. Using matching individual-level genotype and phenotype data for 27 traits in the Estonian biobank 13 and genotype data directly from the ancient source populations, we quantify the contributions from each ancestry to present-day phenotypic variation in each complex trait. We find substantial differences in ancestry for eye and hair color, body mass index, waist/hip circumferences, and their ratio, height, cholesterol levels, caffeine intake, heart rate, and age at menarche. Furthermore, we find evidence for recent positive selection linked to four of these traits and, in addition, sleep patterns and blood pressure. Our results show that these ancient components were differentiated enough to contribute ancestry-specific signatures to the complex trait variability displayed by contemporary Europeans.This is a fascinating effort, but I'm not taking it too seriously until I see the results reproduced with several cohorts from very different parts of Europe. The reason being is that at least some of the outcomes might be specific to Estonia, and reflective of its own peculiar recent population history. For example, the authors find that among Estonians blond hair and blue eyes show a high association with Anatolian farmer ancestry (see table S4). Now, some people might be surprised by this link between light pigmentation and Near Eastern ancestry. However, I'm not, because I know that quite a few Estonians, especially northwest Estonians, harbor recent north German and/or Scandinavian ancestry. Obviously, north Germans and Scandinavians are some of the blondest haired and lightest eyed people in Europe. But they also have more Anatolian farmer ancestry than Estonians. So it might well be that in Estonia these traits are strongly linked with recent Germanic ancestry rather than ancient Anatolian ancestry. In fact I'm willing to bet that this is indeed the case. I'm also willing to bet that blond hair and blue eyes won't show a strong association with Anatolian farmer ancestry in other European countries, but rather with steppe herder ancestry or even, in some cases, minor Siberian admixture. Citation... Marnetto et al., Ancestral genomic contributions to complex traits in contemporary Europeans, Current Biology (2022), https://doi.org/10.1016/j.cub.2022.01.046 See also... Mainstream media: Europeans owe their height to Asian nomads Blond hair is only indirectly associated with Anatolian ancestry in Estonia...duh
Wednesday, February 2, 2022
The PIE homeland controversy: February 2022 status report
I think we'll see the emergence of two main competing proto-Indo-European (PIE) homeland theories over the next few years:
- a homeland in the Eneolithic North Caucasus, and the spread of Anatolian languages into West Asia with Maykop-related ancestry - a homeland in the North Pontic region, possibly within the Eneolithic Sredny Stog archeological culture, and the spread of Anatolian languages into West Asia via the Balkans.Both theories have support from ancient DNA. Some of it has already been published (for instance, see here). At this point, I can see myself firmly in the North Pontic camp, even if it turns out that North Pontic-related ancestry only made a fleeting impact on Bronze Age Anatolia. After all, there's no direct relationship between genes and languages, so to prove that Anatolian languages came from the North Pontic, there's no need for North Pontic-related ancestry to persist in Anatolia, as long as we have solid evidence that people with this type of ancestry moved there at the right time. In my mind, for now, the Maykop culture provides an excellent explanation for non-Indo-European influences in PIE, and there's no need to make it Indo-European speaking, let alone PIE speaking. See also... The PIE homeland controversy: June 2021 status report
Sunday, January 23, 2022
Para-Turbo-Balto-Slavic?
I'm seeing increasing numbers of Bronze and Iron Age samples from Central Europe and surrounds with this peculiar set of traits:
- shared genetic drift with present-day Balto-Slavic speakers to the exclusion of most other Europeans - and yet, an unusually low level of Yamnaya-related steppe ancestry - so much so, in fact, that they're often outside the range of modern European genetic variation.As far as I can tell, currently the best examples of this unusual population are HUN_Mako_EBA_o:I1502 (Mathieson et al. Nature 2015) and HUN_EIA_Prescythian_Mezocsat_o1:I18241 (Patterson et al. Nature 2021). Both are from the Carpathian Basin in what is now Hungary. I ran a series of qpAdm mixture models to try and learn more about their origins. The most robust outcomes, out of about 50 different attempts, are these:
right pops: CMR_Shum_Laka_8000BP MAR_Taforalt IRN_Ganj_Dareh_N Levant_PPNB TUR_Barcin_N Iberia_Southeast_Meso UKR_Meso England_Meso RUS_Karelia_HG RUS_West_Siberia_HG MNG_North_N TWN_Hanben BRA_LapaDoSanto_9600BP HUN_Mako_EBA_o Baltic_LTU_Narva 0.149 ∓0.028 POL_Globular_Amphora 0.613 ∓0.028 Yamnaya_RUS_Samara 0.238 ∓0.029 chisq 10.836 tail prob 0.370463 Full output HUN_EIA_Prescythian_Mezocsat_o1 Baltic_LTU_Narva 0.186 ∓0.028 POL_Globular_Amphora 0.592 ∓0.027 Yamnaya_RUS_Samara 0.222 ∓0.029 chisq 12.492 tail prob 0.253499 Full outputCombining the two genomes produces a very similar result:
HUN_EBA-EIA_o Baltic_LTU_Narva 0.160 ∓0.023 POL_Globular_Amphora 0.612 ∓0.023 Yamnaya_RUS_Samara 0.227 ∓0.023 chisq 14.653 tail prob 0.14524 Full outputImportantly, when I move RUS_Karelia_HG from the right pops to the left pops, to test whether HUN_EBA-EIA_o really has steppe ancestry, as opposed to closely related hunter-gatherer ancestry, I still get a very similar outcome:
HUN_EBA-EIA_o Baltic_LTU_Narva 0.158 ∓0.027 POL_Globular_Amphora 0.605 ∓0.033 RUS_Karelia_HG 0.014 ∓0.038 Yamnaya_RUS_Samara 0.223 ∓0.053 chisq 10.461 tail prob 0.234171 Full outputSo these largely Globular Amphora-related individuals do harbor as much as a quarter of steppe ancestry, which is to be expected considering the massive genetic turn-over that most of Europe experienced just before their time as a result of population expansions from the Pontic-Caspian steppe. Nevertheless, this is ~20% less steppe ancestry than in the present-day populations of the region, and it clearly shows in any decent Principal Component Analysis (PCA) of West Eurasia. For instance: At the same time, the relatively close genetic relationship between these ancients and present-day Balto-Slavic speaking populations shows up in fine-scale intra-European PCA. The origins and implications of this population are still a mystery to me. I don't think it's native to the Carpathian Basin. Indeed, my qpAdm models suggest that it may have moved into this region from somewhere to the northeast, because its ancestry is best modeled with ancient groups from present-day Lithuania, Poland and Russia. I'm adamant that these people weren't Balto-Slavic speakers, and certainly not proto-Slavs. Rather, I suspect that much like the Welzin warriors of Bronze Age North-Central Europe, they were closely related to a contemporaneous group that eventually gave rise to proto-Slavs. At best, they may have somehow contributed to the ethnogenesis of Balto-Slavs. By the way, using the Global25 to model their ancestry is highly problematic, because of the strong Balto-Slavic genetic drift that affects some of the dimensions. So be careful when you try it, or better yet, don't try it at all, and stick to formal stats in this particular instance. See also... Tollense Valley Bronze Age warriors were very close relatives of modern-day Slavs
Friday, January 21, 2022
Yamnaya is from Europe, but it's really from Asia
I was about to post a comment under a new preprint at bioRxiv, but the comment section isn't there anymore. Hopefully, this is just a temporary glitch.
The preprint in question is titled Reconstructing the spatiotemporal patterns of admixture during the European Holocene using a novel genomic dating method [LINK]. It's co-authored by Harvard/Broad MIT scientist Nick Patterson who occasionally comments at this blog.
My impression is that the authors see the people associated with the Yamnaya culture as Asians who simply used "far" Eastern Europe as a springboard to expand into other parts of Europe.
If so, they're dead wrong.
There are at least three arguments why the Yamnaya population should be seen as quintessentially European:
- its home was initially and overwhelmingly the Pontic-Caspian steppe, which is entirely located within the present-day borders of Europe - Yamnaya genomes are clearly different from those of older populations native to nearby parts of Asia, and, in fact, these differences show a very strong correlation with the present-day borders between Europe and Asia - the Yamnaya people weren't a new population in Europe by any stretch, but must have been overwhelmingly derived from the very similar Eneolithic peoples of the Pontic-Caspian steppe and/or the nearby forest steppe, both of which are located in Eastern Europe.And yet, this is what the preprint claims:
The beginning of the Bronze Age was a period of major cultural and demographic change in Eurasia, accompanied by the spread of Yamnaya Steppe Pastoralist-related ancestry from Pontic-Caspian steppes into Europe and South Asia (16).In fact, what really happened at this time was that Yamnaya steppe pastoralist-related ancestry spread from Eastern Europe to other parts of Europe, as well as to Central and West Asia. The preprint does eventually explain that present-day South Asians derive their Yamnaya-related ancestry from a later eastward expansion of the European Corded Ware culture (CWC), but it completely ignores the fact that the Afanasievo culture was the result of the initial eastward expansion from Europe to Asia. That is, the ancestors of the Afanasievo people were recent migrants from the Pontic-Caspian steppe to Central Asia and Siberia. There's also this:
Over the following millennium, the Yamnaya-derived groups of the Corded Ware Complex (CWC) and Bell Beaker complex (BBC) cultures brought Steppe pastoralist-related ancestry to Europe.Seriously? Both the CWC and BBC, just like the Yamnaya culture, were from Europe. In fact, as per above, the descendants of the CWC expanded into Asia. And this:
The second major migration occurred when populations associated with the Yamnaya culture in the Pontic-Caspian steppe expanded to central and western Europe from far eastern Europe.The authors basically admit here that Yamnaya came from Eastern Europe, but they call it "far" Eastern Europe. Perhaps they know something I don't, but as things stand, there's no evidence that Yamnaya came from "far" Eastern Europe. In fact, the emerging consensus based on ancient DNA, including pre-publication data, is that Yamnaya may have originated in what is now Ukraine. In my opinion, Ukraine isn't located in "far" Eastern Europe, but more or less in the middle of it. Inexplicably, this is what they say about the genetic origins of the Yamnaya and Afanasievo peoples:
These groups were likely the result of a genetic admixture between the descendants of EHG-related groups and CHG-related groups associated with the first farmers from Iran (8, 22, 36). ... Thus, we combined all early Steppe pastoralist individuals in one group to obtain a more precise estimate for the genetic formation of proto-Yamnaya of ~4,400 to 4,000 BCE (Figure 2). These dates are noteworthy as they pre-date the archeological evidence by more than a millennium (37) and have important implications for understanding the origin of proto-Pontic Caspian cultures and their spread to Europe and South Asia.Not really. Like I said, the Yamnaya population was overwhelmingly derived from the Eneolithic peoples of the Eastern European steppe and/or forest steppe. And these Yamnaya-like Eneolithic peoples were spread out across a vast area of Eastern Europe by at least ~4,500 BCE. Some of their genomes have been available for several years, and many more are on the way. It is possible that the Yamnaya and Afanasievo genotype formed in 4,400-4,000 BCE, but if so, then this was due to mixing between the Eneolithic steppe peoples and nearby European farmers. That's because the difference between the Yamnaya and Eneolithic steppe genotypes is minor (~15%) European farmer admixture in the former. The really interesting puzzle is exactly where and when the peculiar Eneolithic steppe genotype came into being. Any ideas Dr Patterson? See also... Matters of geography Understanding the Eneolithic steppe
Labels:
Afanasevo,
Afanasievo,
ancient DNA,
Corded Ware,
Corded Ware Culture,
CWC,
Eastern Europe,
Eurasia,
Eurasian steppe,
Indo-European,
Pontic-Caspian steppe,
Proto-Indo-European,
Yamna,
Yamnaya
Tuesday, January 18, 2022
Mistaken identity?
Ancient Bohemian I20509 is dated to 400-200 BCE, or the La Tene period, in Patterson et al. 2021 (see here). However, he belongs to Y-chromosome N-L550 and is most similar to northern Swedes in my Global25 analysis. So I reckon he's a Swedish soldier who may have died during the Thirty Years' War. In any case, he seems to be a lot younger than the La Tene period, so, for now, I've labeled him CZE_IA_La_Tene_oFennoscandian in the Global25 datasheets (see here).
See also...
They came, they saw, and they mixed
Subscribe to:
Posts (Atom)