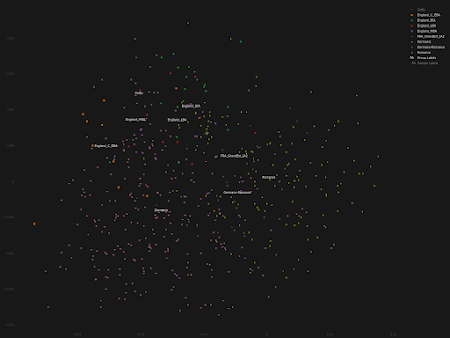
- significant population shifts need not result in any noticeable changes in ancient ancestry proportions - ancient ancestry proportions can shift without significant migrations from afar due to cryptic population substructures - large-scale population shifts need not result in langage shifts, especially if they're gradual - small-scale population shifts can result in language shifts, especially if they're sudden.Indeed, when I plot some of the key ancient samples from the paper in my ultra fine scale Principal Component Analyses (PCA) of Northern and Western Europe, it appears that it's only the Early Iron Age (EIA) population from England that overlaps significantly with a roughly contemporaneous group from nearby Celtic-speaking continental Europe. The relevant PCA data are available here and here, respectively. See also... Celtic vs Germanic Europe Avalon vs Valhalla revisited R1a vs R1b in third millennium BCE central Europe
search this blog
Thursday, December 23, 2021
When did Celtic languages arrive in Britain?
A new paper at Nature by Patterson et al. argues that Celtic languages spread into Britain during the Bronze Age rather than the Iron Age [LINK]. This argument is based on the observation that there was a large-scale shift in deep ancestry proportions in Britain during the Bronze Age.
In particular, the ratio of Early European Farmer (EEF) ancestry increased significantly in what is now England during the Late Bronze Age (LBA). On the other hand, the English Iron Age was a much more stable period in this context.
I don't have any strong opinions about the spread of Celtic languages into Britain, and Patterson et al. might well be correct, but their argument is potentially flawed because:
Tuesday, November 9, 2021
Crazy stuff
I'm hoping that 2022 is the year when this problem is finally straightened out. Over to you David Reich, Nick Patterson, Iosif Lazaridis, David Anthony, Wolfgang Haak, Johannes Krause and colleagues.
See also...
An early Iranian, obviously
The Hajji Firuz fiasco
A Mycenaean and an Iron Age Iranian walk into a bar...
Wednesday, October 27, 2021
Local origins of the earliest Tarim Basin mummies (Zhang et al. 2021)
Over at Nature at this LINK. It's nice to see yet another huge surprise courtesy of ancient DNA. Please note that most of the ancients from this paper are already in the Global25 datasheets. Here's the abstract:
The identity of the earliest inhabitants of Xinjiang, in the heart of Inner Asia, and the languages that they spoke have long been debated and remain contentious 1. Here we present genomic data from 5 individuals dating to around 3000–2800 bc from the Dzungarian Basin and 13 individuals dating to around 2100–1700 bc from the Tarim Basin, representing the earliest yet discovered human remains from North and South Xinjiang, respectively. We find that the Early Bronze Age Dzungarian individuals exhibit a predominantly Afanasievo ancestry with an additional local contribution, and the Early–Middle Bronze Age Tarim individuals contain only a local ancestry. The Tarim individuals from the site of Xiaohe further exhibit strong evidence of milk proteins in their dental calculus, indicating a reliance on dairy pastoralism at the site since its founding. Our results do not support previous hypotheses for the origin of the Tarim mummies, who were argued to be Proto-Tocharian-speaking pastoralists descended from the Afanasievo 1,2 or to have originated among the Bactria–Margiana Archaeological Complex 3 or Inner Asian Mountain Corridor cultures 4. Instead, although Tocharian may have been plausibly introduced to the Dzungarian Basin by Afanasievo migrants during the Early Bronze Age, we find that the earliest Tarim Basin cultures appear to have arisen from a genetically isolated local population that adopted neighbouring pastoralist and agriculturalist practices, which allowed them to settle and thrive along the shifting riverine oases of the Taklamakan Desert.Zhang, F., Ning, C., Scott, A. et al. The genomic origins of the Bronze Age Tarim Basin mummies. Nature (2021). https://doi.org/10.1038/s41586-021-04052-7 See also... How the Shirenzigou nomads became Proto-Tocharians
Wednesday, October 20, 2021
Modern domestic horses came from the Eastern European steppe
Over at Nature at this LINK. I'm getting the impression that geneticists and the editors at Nature are really crap at geography. Obviously, this paper argues that modern domestic horses came from the Pontic-Caspian steppe, which is located very firmly in Eastern Europe. But, inexplicably, instead of actually saying this, the authors came up with the much more ambiguous term Western Eurasian steppes, and even put that in the title. I wonder why? Here's the paper abstract:
Domestication of horses fundamentally transformed long-range mobility and warfare 1. However, modern domesticated breeds do not descend from the earliest domestic horse lineage associated with archaeological evidence of bridling, milking and corralling 2,3,4 at Botai, Central Asia around 3500 bc3. Other longstanding candidate regions for horse domestication, such as Iberia 5 and Anatolia 6, have also recently been challenged. Thus, the genetic, geographic and temporal origins of modern domestic horses have remained unknown. Here we pinpoint the Western Eurasian steppes [my note: they actually mean the Pontic-Caspian steppe, which is located in Eastern Europe], especially the lower Volga-Don region, as the homeland of modern domestic horses. Furthermore, we map the population changes accompanying domestication from 273 ancient horse genomes. This reveals that modern domestic horses ultimately replaced almost all other local populations as they expanded rapidly across Eurasia from about 2000 bc, synchronously with equestrian material culture, including Sintashta spoke-wheeled chariots. We find that equestrianism involved strong selection for critical locomotor and behavioural adaptations at the GSDMC and ZFPM1 genes. Our results reject the commonly held association 7 between horseback riding and the massive expansion of Yamnaya steppe pastoralists into Europe [my note: the Yamnaya culture was located in Europe] around 3000 bc 8,9 driving the spread of Indo-European languages 10. This contrasts with the scenario in Asia where Indo-Iranian languages, chariots and horses spread together, following the early second millennium bc Sintashta culture 11,12.Librado, P., Khan, N., Fages, A. et al. The origins and spread of domestic horses from the Western Eurasian steppes. Nature (2021). https://doi.org/10.1038/s41586-021-04018-9 Update: I emailed one of the lead authors, Ludovic Orlando, asking him for a comment. Here it is:
Thanks for your interest in our research. We indeed struggled finding the term that would be most appropriate and this was discussed with our coauthors. The Pontic-Caspian steppe would seem the most obvious choice but my understanding is that this would include a large region, stretching from the most north-western side of the Black sea to the foothills of the Urals. This is larger than the signature recovered in our data. My understanding is that the Eastern European steppes would also stretch more northernly than the region that we narrowed down. Eastern European steppes was also not immediately clear, even for European scholars such as myself. Therefore, it did not seem that there were any terms that were ready-made for truly qualifying our findings. We thus went for Western Eurasian steppes in the main title, and sticked to more precise locations such as the Don-Volga region in the main text. I guess that this is one of those cases where the activities of past herders did not exactly follow some geographic terms that would only be defined thousands of years later.However, the Pontic-Caspian steppe and the Eastern European steppe are in fact terms that describe the western end of the Eurasian steppe. So they should be totally interchangeable with the term Western Eurasian steppes. Except, at least to me, they seem less ambiguous. Ergo, the Eastern European steppe can't be more northerly than the Western Eurasian steppes, because it's the same thing. Moreover, the Pontic-Caspian steppe can't stretch further west than the Western Eurasian steppes, because, again, it's the same thing. Indeed, the land north of the Eastern European/Western Eurasian steppes is called the forest steppe. See also...
Friday, October 15, 2021
Coming soon?
This ISBA9 abstract seems to be highly relevant to the ultimate origins of the Yamnaya and Corded Ware peoples. Emphasis is mine:
Genomic signals of continuity and admixture in the Caucasus Ghalichi Ayshin et al. Situated between the Black and Caspian Sea, the Caucasus is a key geographic region that connects the Near East and the Eurasian Steppe, with a great ecological diversity of ecotones and landscapes rich in natural resources. A recent archaeogenetic study has shown that the genetically diverse Eneolithic and Bronze Age groups of the steppe and mountains correspond to eco-geographic zones in the Caucasus. However, the formation, interactions and population dynamics warrant further investigation. In this study we explore new genome-wide data of 68 individuals from 20 archaeological cultures across the Caucasus mountains, the piedmont and the steppe extending our temporal transect to 6000 years, doubling the number of available genomes from the region. We present the first genomic data from a Mesolithic individual (6100 calBCE) from the Northwest Caucasus that shows Eastern hunter-gatherer ancestry, Neolithic individuals from Georgia, as well as new data from genetically unexplored regions/cultures in the northeastern highlands and the dry steppe. We observe a degree of genetic continuity through time within the main mountain and steppe genetic groups, but also identify various episodes of gene flow between these and the neighboring regions. In the Late Eneolithic period, we find evidence of admixture from the south into the steppe groups, detectable through the presence of Anatolian_Neolithic-like ancestry. During the Bronze Age, we found in Steppe Maykop individuals a genetic link to West Siberian hunter-gatherers, a component that is absent from Yamnaya, North Caucasus and Catacomb groups, but reappears in Bronze Age individuals associated with the Lola culture.I'm not quite sure what it's saying though. Is the Mesolithc individual from the Northwest Caucasus actually an Eastern European hunter-gatherer, or, as I'm expecting, a mixture between Caucasus and Eastern European hunter-gatherers? If the latter, then it's game over for the Out-of-Iran and Out-of-Armenia Indo-European hypotheses that have been so popular among academics in recent years. The authors also mention the spread of Anatolian-related ancestry into the Eastern European steppe during the Late Eneolithic. They're probably referring to the phenomenon that gave rise to the so called Steppe Maykop outliers. The ISBA9 abstract PDF book is freely available here. See also... Understanding the Eneolithic steppe Ancient DNA vs Ex Oriente Lux A note on Steppe Maykop
Monday, September 27, 2021
The genetic origin and legacy of the Etruscans (Posth et al. 2021)
Over at Science Advances at ths LINK. I'll take a closer look at this issue after I get the relevant genotype data. Anyone got the link? Here's the paper abstract:
The origin, development, and legacy of the enigmatic Etruscan civilization from the central region of the Italian peninsula known as Etruria have been debated for centuries. Here we report a genomic time transect of 82 individuals spanning almost two millennia (800 BCE to 1000 CE) across Etruria and southern Italy. During the Iron Age, we detect a component of Indo-European–associated steppe ancestry and the lack of recent Anatolian-related admixture among the putative non–Indo-European–speaking Etruscans. Despite comprising diverse individuals of central European, northern African, and Near Eastern ancestry, the local gene pool is largely maintained across the first millennium BCE. This drastically changes during the Roman Imperial period where we report an abrupt population-wide shift to ~50% admixture with eastern Mediterranean ancestry. Last, we identify northern European components appearing in central Italy during the Early Middle Ages, which thus formed the genetic landscape of present-day Italian populations.Citation: C. Posth, V. Zaro, M. A. Spyrou, S. Vai, G. A. Gnecchi-Ruscone, A. Modi, A. Peltzer, A. Mötsch, K. Nägele, &. J. Vågene, E. A. Nelson, R. Radzevičiūtė, C. Freund, L. M. Bondioli, L. Cappuccini, H. Frenzel, E. Pacciani, F. Boschin, G. Capecchi, I. Martini, A. Moroni, S. Ricci, A. Sperduti, M. A. Turchetti, A. Riga, M. Zavattaro, A. Zifferero, H. O. Heyne, E. Fernández-Domínguez, G. J. Kroonen, M. McCormick, W. Haak, M. Lari, G. Barbujani, L. Bondioli, K. I. Bos, D. Caramelli, J. Krause, The origin and legacy of the Etruscans through a 2000-year archeogenomic time transect. Sci. Adv. 7, eabi7673 (2021). See also... Etruscans, Latins, Romans and others
Wednesday, September 15, 2021
Yamnaya people drank horse milk (Wilkin et al. 2021)
Over at Nature at this LINK. I'm guessing the claim that Yamnaya pastoralists lived in Scandinavia is a huge typo. Obviously, the authors are referring to the people of the Corded Ware culture (CWC). From the paper:
During the Early Bronze Age, populations of the western Eurasian steppe expanded across an immense area of northern Eurasia. Combined archaeological and genetic evidence supports widespread Early Bronze Age population movements out of the Pontic–Caspian steppe that resulted in gene flow across vast distances, linking populations of Yamnaya pastoralists in Scandinavia with pastoral populations (known as the Afanasievo) far to the east in the Altai Mountains1,2 and Mongolia3. Although some models hold that this expansion was the outcome of a newly mobile pastoral economy characterized by horse traction, bulk wagon transport4,5,6 and regular dietary dependence on meat and milk5, hard evidence for these economic features has not been found. Here we draw on proteomic analysis of dental calculus from individuals from the western Eurasian steppe to demonstrate a major transition in dairying at the start of the Bronze Age. The rapid onset of ubiquitous dairying at a point in time when steppe populations are known to have begun dispersing offers critical insight into a key catalyst of steppe mobility. The identification of horse milk proteins also indicates horse domestication by the Early Bronze Age, which provides support for its role in steppe dispersals. Our results point to a potential epicentre for horse domestication in the Pontic–Caspian steppe by the third millennium bc, and offer strong support for the notion that the novel exploitation of secondary animal products was a key driver of the expansions of Eurasian steppe pastoralists by the Early Bronze Age.Wilkin, S., Ventresca Miller, A., Fernandes, R. et al. Dairying enabled Early Bronze Age Yamnaya steppe expansions. Nature (2021). https://doi.org/10.1038/s41586-021-03798-4 See also... On the origin of the Corded Ware people
Saturday, September 4, 2021
The genomic formation of modern Balkan peoples (Olalde et al. 2021 preprint)
Over at bioRxiv at this LINK. This preprint deals with some very complex issues, so I can't say much about it until I have a good look at the relevant genotype data. However, for now, my impression is that the authors have oversimplified the genetic origins of most Balkan peoples.
For instance, they model the present-day Greek population as a two way mixture between ancient Greeks from a Greek colony in Iberia and present-day Mordovians. The Mordovians are basically a proxy for the Slavs who moved into the Balkans during the Medieval period.
However, the problem is that, strictly speaking, this isn't a historically plausible model, because Mordovians are actually a Uralic-speaking group from the Volga region with significant Siberian ancestry. Needless to say, it's extremely unlikely that anyone like them had an appreciable impact on the present-day Greek gene pool.
So instead I'd like to see the authors try three-way and four-way models with ancients from Mycenae, Anatolia and some places (well to the west of the Volga River) likely to have been inhabited by early Slavs.
Feel free to let me know what you think about this preprint in the comments below. Here's the abstract:
The Roman Empire expanded through the Mediterranean shores and brought human mobility and cosmopolitanism across this inland sea to an unprecedented scale. However, if this was also common at the Empire frontiers remains undetermined. The Balkans and Danube River were of strategic importance for the Romans acting as an East-West connection and as a defense line against “barbarian” tribes. We generated genome-wide data from 70 ancient individuals from present-day Serbia dated to the first millennium CE; including Viminacium, capital of Moesia Superior province. Our analyses reveal large scale-movements from Anatolia during Imperial rule, similar to the pattern observed in Rome, and cases of individual mobility from as far as East Africa. Between ∼250-500 CE, we detect gene-flow from Central/Northern Europe harboring admixtures of Iron Age steppe groups. Tenth-century CE individuals harbored North-Eastern European-related ancestry likely associated to Slavic-speakers, which contributed >20% of the ancestry of today’s Balkan people.Olalde et al., Cosmopolitanism at the Roman Danubian Frontier, Slavic Migrations, and the Genomic Formation of Modern Balkan Peoples, bioRxiv, posted August 31, 2021, doi: https://doi.org/10.1101/2021.08.30.458211 See also... A Greek tragedy
Friday, August 27, 2021
R1a vs R1b in third millennium BCE Central Europe (Papac et al. 2021)
R1a-M417 and R1b-L51 are by far the most important Y-chromosome haplogroups in Europe today. More precisely, R1a-M417 dominates in Eastern Europe, while R1b-L51 in Western Europe.
It's been obvious for a while now, at least to me, that both of these Y-haplogroups are closely associated with the men of the Late Neolithic Corded Ware culture (CWC). Indeed, in my mind they're the main genetic signals of its massive expansion, probably from a homeland somewhere north of the Black Sea in what is now Ukraine.
I'm still not exactly sure how the east/west dichotomy between R1a and R1b emerged in Europe, but, thanks to a new paper by Papac et al. at Science Advances, at least now I have a working hypothesis about that. Below is a quote from the said paper, emphasis is mine:
In addition to autosomal genetic changes through time, we observe a sharp reduction in Y-chromosomal diversity going from five different lineages in early CW to a dominant (single) lineage in late CW (Fig. 4A). We used forward simulations to explore the demographic scenarios that could account for the observed reduction in Y-chromosomal diversity. Performing 1 million simulations of a population with a starting frequency of R1a-M417(xZ645) centered around the observed starting frequency in Bohemia_CW_Early (3 of 11, 0.27), we assessed the plausibility of this lineage reaching the observed frequency in Bohemia_CW_Late (10 of 11, 0.91) in the time frame of 500 years under a model of a closed population and random mating (Materials and Methods). We reject the “neutral” hypothesis, i.e., that this change in frequency occurred by chance, given a wide range of plausible population sizes. Instead, our results suggest that R1a-M417(xZ645) was subject to a nonrandom increase in frequency, resulting in these males having 15.79% (4.12 to 44.42%) more surviving offspring per generation relative to males of other Y-haplogroups. We also find that this change in Y chromosome frequency is extreme compared to the changes in allele frequencies at fully covered autosomal 1240k sites within the same males, suggesting a process that disproportionately affected Y-chromosomal compared to autosomal genetic diversity, ruling out a population bottleneck as the likely cause. Our results suggest that the Y-lineage diversity in early CW males was supplanted by a nonrandom process [selection, social structure, or influx of nonlocal R1a-M417(xZ645) lineages] that drove the collapse in Y-chromosomal diversity. A simultaneous decline of Y-chromosomal diversity dating to the Neolithic has been observed across most extant Y-haplogroups (64), possibly due to increased conflict between male-mediated patrilines (65). We view that changes in social structure (e.g., an isolated mating network with strictly exclusive social norms) could be an alternative cause but would be difficult to distinguish in the underlying model parameters.Right, so even though the CWC was clearly a community of closely related groups, there must have been some competition between its different clans. And since these clans were highly patriarchal and patrilineal, this competition probably led to different paternal lineages dominating different parts of the CWC horizon, with M417 becoming especially common in the east and L51 in the west. Of course, the expansions of post-Corded Ware groups, such as the M417-rich Slavs in Eastern Europe and L51-rich Celts in Western Europe, were also instrumental in creating Europe's R1a/R1b dichotomy, but obviously these groups were in large part the heirs of the CWC. By the way, most of the samples from Papac et al. are already in the Global25 datasheets linked here. Look for the labels listed here. Below is a plot made from the Global25 data courtesy of regular commentator Matt. Citation: L. Papac, M. Ernée, M. Dobeš, M. Langová, A. B. Rohrlach, F. Aron, G. U. Neumann, M. A. Spyrou, N. Rohland, P. Velemínský, M. Kuna, H. Brzobohatá, B. Culleton, D. Daněček, A. Danielisová, M. Dobisíková, J. Hložek, D. J. Kennett, J. Klementová, M. Kostka, P. Krištuf, M. Kuchařík, J. K. Hlavová, P. Limburský, D. Malyková, L. Mattiello, M. Pecinovská, K. Petriščáková, E. Průchová, P. Stránská, L. Smejtek, J. Špaček, R. Šumberová, O. Švejcar, M. Trefný, M. Vávra, J. Kolář, V. Heyd, J. Krause, R. Pinhasi, D. Reich, S. Schiffels, W. Haak, Dynamic changes in genomic and social structures in third millennium BCE central Europe. Sci. Adv. 7, eabi6941 (2021). See also... On the origin of the Corded Ware people Understanding the Eneolithic steppe Conan the Barbarian probably belonged to Y-haplogroup R1a
Labels:
ancient DNA,
Balto-Slavic,
Celtic,
Central Europe,
Corded Ware,
Corded Ware Culture,
CWC,
Eastern Europe,
Germanic,
Indo-European,
Northern Europe,
Proto-Indo-European,
R1a-M417,
R1b-L151,
R1b-L51,
Slavic,
Slavs
Tuesday, July 20, 2021
On the origin of the Corded Ware people
There's been a lot of talk lately about the finding that the peoples associated with the Corded Ware and Yamnaya archeological cultures were genetic cousins (for instance, see here). As I've already pointed out, this is an interesting discovery, but, at this stage, it's difficult to know what it means exactly.
It might mean that the Yamnayans were the direct predecessors of the Corded Ware people. Or it might just mean that, at some point, the Corded Ware and Yamnaya populations swapped women regularly (that is, they practiced female exogamy with each other).
In any case, I feel that several important facts aren't being taken into account by most of the interested parties. These facts include, in no particular order:
- despite being closely related, the Corded Ware and Yamnaya peoples were highly adapted to very different ecological zones - temperate forests and arid steppes, respectively - and this is surely not something that happened within a few years and probably not even within a couple of generations - both the Corded Ware and Yamnaya populations expanded widely and rapidly at around the same time, but never got in each others way, probably because they occupied very different ecological niches - despite sharing the R1b Y-chromosome haplogroup, their paternal origins were quite different, with Corded Ware males rich in R1a-M417 and R1b-L51 and Yamnaya males rich in R1b-Z2103 and I2a-L699I suppose it's possible that the Corded Ware people were overwhelmingly and directly derived from the Yamnaya population. But right now my view is that, even if they were, then the Yamnaya population that they came from was quite different from the classic, R1b-Z2103-rich Yamnaya that spread rapidly across the steppes. Indeed, perhaps what we're dealing with here is a very early (proto?) Yamnaya gene pool located somewhere in the border zone between the forests and the steppes, that then split into two main sub-populations, with one of these groups heading north and the other south? I do wonder what David Anthony would say if he was made aware of the above mentioned facts? Then again, perhaps he's already aware of them, and simply chose to ignore them when formulating his latest theory about the origin of the Corded Ware people? See also...
Monday, June 28, 2021
The PIE homeland controversy: June 2021 status report
Archeologist David Anthony has made several appearances online recently to promote his theories about the origins of the Corded Ware and Yamnaya cultures and peoples.
In a clip on Youtube he reiterated his theory that the so called Iranian-related ancestry in the Yamnaya people actually came from what is now Iran, and, more precisely, that it was carried by hunter-gatherers who travelled relatively rapidly from the South Caspian region into the Volga Delta in what is now Russia.
It's still a complete mystery to me as to why a group of hunter-gatherers from the South Caspian would undertake such a migration, instead of, say, expanding their range gradually over thousands of years, first into the Caucasus and eventually into Eastern Europe.
But there's a more serious problem with Anthony's theory: it contradicts the currently available ancient DNA. That's because the so called Iranian-related ancestry in the Yamnaya people is most closely related to the Kotias and Satsurblia hunter-gatherers from what is now Georgia, and these hunter-gatherers form a separate clade from the earliest samples from what is now Iran. For instance, see here and here.
Also, in a podcast on Razib's blog, Anthony doubled down on his theory that Y-chromosome haplogroup R1a was closely associated with Yamnaya plebs who were excluded from Kurgan burials, and, as a result, their remains haven't yet been sampled.
At least this theory isn't yet contradicted by ancient DNA, but it's more complicated and less parsimonious than my theory, which posits that R1a, or rather R1a-M417, was simply a very rare lineage in the Yamnaya population, and that it only became a common and widespread marker thanks to the Corded Ware expansion (see here).
Intriguingly, my understanding is that there are several unpublished R1a samples from the Caspian and Volga steppes at Harvard's David Reich Lab that have been classified by its scientists as Yamnaya outliers. Of course, Anthony is collaborating on at least one major paper with this lab (see here).
Ergo, I strongly suspect that Anthony's theory is in part based on these Yamnaya outliers. However, I also believe that these samples are wrongly dated and probably represent Scythians and/or Sarmatians. I'll be able to look into that if they're ever published.
Speaking of the David Reich Lab, its leading scientists, David Reich and Nick Patterson, have also made appearances online recently, on Youtube and Razib's blog, respectively, to reveal that the Corded Ware and Yamnaya peoples aren't just very similar genetically, but in fact close cousins.
This is a very interesting finding. Apparently it's based on a relatively high level of Identity-by-Descent (IBD) segment sharing between Corded Ware and Yamnaya samples, but that's all I know. I'm guessing that the relevant paper is coming soon (that is, within the next five years).
However, the long-standing question that the readers of this blog want to see answered is not whether the Corded Ware and Yamnaya peoples are close cousins, but whether Yamnaya migrants founded the Corded Ware culture. The obvious way to prove that they did is to find at least one ancient population unambiguously classified as part of the Yamnaya horizon that is rich in the typically Corded Ware Y-haplogroups R1a-M417 and R1b-L151.
See also...
On the origin of the Corded Ware people
The PIE homeland controversy: January 2019 status report
The PIE homeland controversy: August 2019 status report
Labels:
ancient DNA,
Corded Ware,
CWC,
David Anthony,
David Reich,
David Reich Lab,
Eastern Europe,
Indo-European,
Iran,
Nick Patterson,
Proto-Indo-European,
R1a-M417,
R1b-L151,
R1b-L51,
Yamna,
Yamnaya
Thursday, June 17, 2021
Balto-Slavic drift
A few years ago I began using the term "Balto-Slavic genetic drift" to describe the fine-scale genetic signal that is shared by the speakers of Baltic and Slavic languages to the exclusion of Europeans without significant Balto-Slavic ancestry.
As a result, nowadays, many people online use the term "Balto-Slavic drift" when referring to this phenomenon.
The easiest way to prove that Balto-Slavic drift exists is to run a fine-scale Principal Component Analysis (PCA) of European genetic variation with a lot of Balto-Slavic samples in the mix. Indeed, my Global25 PCA analysis does a great job of illustrating the impact of Balto-Slavic drift on the population structure of Europe both in PCA plots and mixture models (for instance, see here).
It's also possible to tease out Balto-Slavic drift with formal statistics. I showed this indirectly in a recent blog post about Greek population structure (see here). In this post I'm going to demonstrate how to explicitly and formally test for Balto-Slavic drift both in ancient and present-day samples.
To do this we need to find stats that basically split Baltic and Slavic speakers from other Europeans, such as f4(Outgroup,Test;Bell_Beaker_NDL,Baltic_LVA_BA). In this f4-stat, Baltic_LVA_BA is the ancient reference population with an unusually high level of Balto-Slavic drift, while Bell_Beaker_NDL is a fairly similar population overall in terms of ancient ancestry components, but with practically zero Balto-Slavic drift.
Note that the statistics with the most significant Z scores (>3) involve populations that speak Baltic or Slavic languages, or their neighbors who plausibly harbor significant Baltic and/or Slavic ancestry. Among the ancient, mostly Scandinavian, populations (from Margaryan et al. 2020 and marked with the VK2020 prefix), significant Balto-Slavic drift only appears in the more easterly and/or later groups from the Viking Age (VA).
Unfortunately, one of the problems with this analysis is that Baltic_LVA_BA and Bell_Beaker_NDL aren't identical in terms of their ancient ancestry proportions. For one, the latter has significantly more Neolithic farmer ancestry. No wonder then, that Greeks, who are mostly of early farmer stock, don't show a significant Z score, despite probably packing a significant amount of Balto-Slavic ancestry dating to the Middle Ages.
In the near future, as more ancient samples become available, it might be possible to find better reference populations for the job and create more accurate, finer-scaled tests.
See also...
Uralian genes
That old chestnut: Northeast vs Northwest Euros
Friday, May 14, 2021
A Greek tragedy
I wasn't going to blog about the Clemente et al. "Aegean palatial civilizations" paper, because I think that it's a rather strange effort overall. But apparently a lot of people want to know my thoughts on the topic, so here goes.
If you download the relevant PDF file (here) and do a search for "Slav", you'll see that the word doesn't even appear in the bibliography. How is that possible, considering the massive impact that the Slavs had on the Balkans, including Greece, during the Middle Ages?
Indeed, here's a quote from page 12 of the PDF: "Present-day Greeks - who also carry Steppe-related ancestry - share ~90% of their ancestry with MBA northern Aegeans, suggesting continuity between the two time periods."
That's a very optimistic view. In fact, there's no evidence whatsoever in the paper that there's even 1% genetic continuity between present-day Greeks and any ancient Greek population, let alone the MBA northern Aegeans.
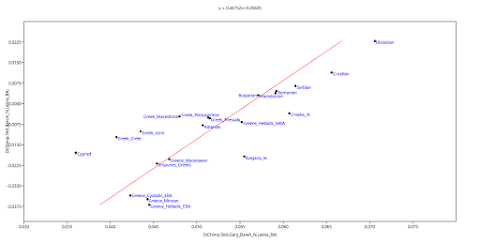
The genetic impact of Medieval Slavic migrations on most present-day Greek populations is easy to see. For instance, below are several linear models based on D-statistics of the form D(Outgroup,Test;Ancient1,Ancient2). You don't need a PhD in mathematics to understand them. The relevant data file is available here.
Note that most of the present-day Greek groups cluster together, and they also form fairly neat clines with the other Greeks, as well as Cypriots, other Balkan populations, including those speaking Slavic languages, and also the Slavic-speaking Ukrainians. On the other hand, they don't overlap with any of the ancient groups from Greece and surrounds, nor do they generally form obvious clines with them.
To me this suggests that most present-day Greeks harbor significant levels of Slavic ancestry and some sort of recent Cypriot-related ancestry, and in large part they're only coincidentally similar to ancient Aegeans, including those from the MBA (labeled Greece_Helladic_MBA in my graphs).
And let me assure you that no matter which ancient populations you run in such D-stats, you'll always see similar present-day Greek clusters and present-day Balkan clines.
Obviously, it's fair enough to assume that there's been some genetic continuity in the Aegean from the Iron Age, Bronze Age, and even the Copper Age and Neolithic era to the present-day. But the point I'm making is that no one has yet proved this, or even attempted to measure it properly.
See also...
Greek confirmation bias
Monday, April 26, 2021
Uralians of the Sargat horizon
Many years ago, well before the start of the ancient DNA revolution, someone made the very clever inference that the N-Tat Y-chromosome marker was closely associated with the expansion of Uralic languages.
Since then, N-Tat has been renamed several times over, to the point that I no longer know what it's called, but the aforementioned inference has turned into a very solid consensus backed up by a wide range of studies focusing on modern and ancient DNA.
Nowadays, Y-haplogroup N-L1026, a subclade of N-Tat, is seen as the main genetic signal of the Uralic expansions, along, of course, with Nganasan-related genome-wide genetic ancestry.
A recent paper at Science Advances by Gnecchi-Ruscone et al. featured the first ever genome-wide samples from the Sargat horizon, which is an Iron Age archeological formation in western Siberia normally associated with the Ugric branch of the Uralic language family. Surprisingly, and disappointingly, the authors failed to investigate this widely accepted connection.
If we go by the Y-haplogroup classifications in the paper, which may or may not be the smart thing to do, at least two of the Sargat horizon males belong to N-L1026, and one also to the more derived N-Z1936 subclade, which has been found in the remains of Hungarian Conquerers from Medieval Hungary. Of course, Hungarian is an Ugric language generally thought to have been introduced into the Carpathian Basin by the Hungarian Conquerers who originally came from western Siberia.
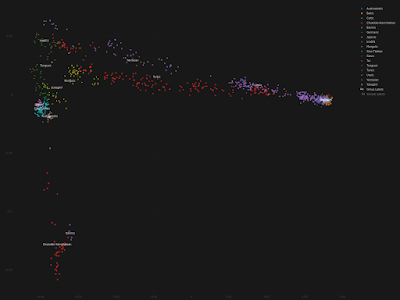
That's probably enough to corroborate the association between the Sargat horizon and the spread of Ugric/Uralic languages, but let's also take a quick look at the autosomal DNA of these Sargat individuals. Firstly, here's a Principal Component Analysis (PCA), based on Global25 data and produced with the Vahaduo G25 Views online tool. The results are self-explanatory.
Interestingly, I can't get a decent statistical fit when I try to reproduce the four-way qpWave/qpAdm model done by Gnecchi-Ruscone et al., probably mostly because my right pops or outgroups are different. This suggests to me that there's something important missing in their model.
Sargat_IA MNG_Khovsgol_LBA 0.203±0.045 RUS_Ekven_IA 0.183±0.044 RUS_Sintashta_MLBA 0.545±0.014 TKM_Gonur1_BA 0.068±0.013 chisq 16.805 tail prob 0.0186971 Full outputSo how about if I replace RUS_Ekven_IA with kra001, the oldest Nganasan-like individual in the ancient DNA record (see here), and MNG_Khovsgol_LBA with KAZ_Mereke_MBA, to add a more local stream of ancestry?
Sargat_IA KAZ_Mereke_MBA 0.135±0.017 kra001 0.301±0.007 RUS_Sintashta_MLBA 0.499±0.023 TKM_Gonur1_BA 0.066±0.015 chisq 8.872 tail prob 0.262001 Full outputThat's a better statistical fit and also, I'd say, a more realistic model, at least in terms of distal ancestry proportions. Note that Nganasan-related ancestry makes up 30% of the genome-wide genetic structure of the Sargat samples, which again corroborates the view that Uralic languages were spoken within the Sargat horizon. Update 28/04/21: This is the best qpAdm model that I could find for Sargat_IA, at least in terms of the chisq and tail prob. It shows that the Sargat population was in large part very similar to that of KAZ_Pazyryk_IA.
Sargat_IA KAZ_Mereke_MBA 0.032±0.016 KAZ_Pazyryk_IA 0.698±0.016 RUS_Sintashta_MLBA 0.236±0.021 TKM_Gonur1_BA 0.034±0.014 chisq 2.023 tail prob 0.958561 Full outputIt's missing kra001, because KAZ_Pazyryk_IA packs enough kra001-related ancestry for the job.
KAZ_Pazyryk_IA KAZ_Mereke_MBA 0.144±0.018 kra001 0.429±0.008 RUS_Sintashta_MLBA 0.378±0.026 TKM_Gonur1_BA 0.049±0.018 chisq 8.899 tail prob 0.259983 Full outputThe fact that KAZ_Pazyryk_IA can be modeled with significant kra001-related ancestry isn't surprising, considering that its territory was located in Siberia. However, my model doesn't necessarily prove that the Sargat population was largely or even partly of Pazyryk origin. Indeed, N-L1026 hasn't yet appeared in any Pazyryk remains. See also... The Uralic cline with kra001 - no projection this time First taste of Early Medieval DNA from the Ural region Hungarian Conquerors were rich in Y-haplogroup N More on the association between Uralic expansions and Y-haplogroup N It was always going to be this way On the association between Uralic expansions and Y-haplogroup N
Labels:
ancient DNA,
Corded Ware,
Indo-European,
Indo-Iranian,
kra001,
N-tat,
N1c,
N1c1a1a,
Nganasan,
Proto-Uralic,
R1a,
Saka,
Sargat,
Siberia,
Sintashta,
Ural,
Uralian,
Uralic
Thursday, April 22, 2021
The history of the Scythians (Gnecchi-Ruscone et al. 2021)
Over at Science Advances at this LINK. Many of the samples from this paper are in the Global25 datasheets. Look for the relevant population and individual IDs from the paper.
The Scythians were a multitude of horse-warrior nomad cultures dwelling in the Eurasian steppe during the first millennium BCE. Because of the lack of first-hand written records, little is known about the origins and relations among the different cultures. To address these questions, we produced genome-wide data for 111 ancient individuals retrieved from 39 archaeological sites from the first millennia BCE and CE across the Central Asian Steppe. We uncovered major admixture events in the Late Bronze Age forming the genetic substratum for two main Iron Age gene-pools emerging around the Altai and the Urals respectively. Their demise was mirrored by new genetic turnovers, linked to the spread of the eastern nomad empires in the first centuries CE. Compared to the high genetic heterogeneity of the past, the homogenization of the present-day Kazakhs gene pool is notable, likely a result of 400 years of strict exogamous social rules.Gnecchi-Ruscone et al. 2021, Ancient genomic time transect from the Central Asian Steppe unravels the history of the Scythians, Science Advances, Sci Adv 7 (13), eabe4414, DOI: 10.1126/sciadv.abe4414 See also... Uralians of the Sargat horizon
Wednesday, April 7, 2021
The Bacho Kiro surprise (Hajdinjak et al. 2021)
Over at Nature at this LINK. The paper focuses on Neanderthal ancestry in Initial Upper Paleolithic (IUP) humans from what is now Bulgaria. But, to me, much more interesting is the claim by its authors that present-day East Asians harbor ancient European, or, at least, European-related ancestry. From the paper, emphasis is mine:
When we explored models of population history that are compatible with the observations above using admixture graphs [28], we found that the IUP Bacho Kiro Cave individuals were related to populations that contributed ancestry to the Tianyuan individual in China as well as, to a lesser extent, to the GoyetQ116-1 and Ust’Ishim individuals (all |Z| < 3; Fig. 2d, Supplementary Information 6). This resolves the previously unclear relationship between the GoyetQ116-1 and Tianyuan individuals [13] without the need for gene flow between these two geographically distant individuals. ... In conclusion, the Bacho Kiro Cave genomes show that several distinct modern human populations existed during the early Upper Palaeolithic in Eurasia. Some of these populations, represented by the Oase1 and Ust’Ishim individuals, show no detectable affinities to later populations, whereas groups related to the IUP Bacho Kiro Cave individuals contributed to later populations with Asian ancestry as well as some western Eurasian humans such as the GoyetQ116-1 individual in Belgium. This is consistent with the fact that IUP archaeological assemblages are found from central and eastern Europe to present-day Mongolia [5,15,16] (Fig. 1), and a putative IUP dispersal that reached from eastern Europe to East Asia. Eventually populations related to the IUP Bacho Kiro Cave individuals disappeared in western Eurasia without leaving a detectable genetic contribution to later populations, as indicated by the fact that later individuals, including BK1653 at Bacho Kiro Cave, were closer to present-day European populations than to present-day Asian populations [29,30].Hajdinjak, M., Mafessoni, F., Skov, L. et al. Initial Upper Palaeolithic humans in Europe had recent Neanderthal ancestry. Nature 592, 253–257 (2021). https://doi.org/10.1038/s41586-021-03335-3 See also... Ust'-Ishim belongs to K-M526
Labels:
admixture,
ancient ancestry,
ancient DNA,
Bacho Kiro Cave,
Chinese,
East Asia,
East Asians,
Eastern Europe,
Eurasia,
Europe,
European,
Ice Age,
Initial Upper Paleolithic,
IUP,
Japanese,
Upper Paleolithic
Wednesday, March 31, 2021
Against the conventional wisdom
I've read some very strange theories over the years trying to explain who was responsible for the so called Caucasus/Iranian-related ancestry in the Yamnaya people.
Proto-Indo-European speaking farmers from what is now Iran? How about Uruk invaders from Mesopotamia? No, wait, they were migrants from India who spoke Sanskrit. Haha.
Nope, it seems that hunter-gatherers rich in this type of ancestry lived north of the Caucasus already during the so called Pottery Neolithic or even the Mesolithic. That's the impression that I'm getting from watching the clip HERE.
This is basically also the idea that I gradually developed at this blog during the last few years, following common sense and logic, but totally against the conventional wisdom in regards to this topic. For instance, see here...
But here's my prediction: Steppe_EMBA only has 10-15% admixture from the post-Mesolithic Near East not including the North Caucasus, and basically all of this comes via female mediated gene flow from farming communities in the Caucasus and perhaps present-day Ukraine.
Modeling Steppe_EMBAOf course, I could've done better with many of the details in my posts, like the dates and archeological links. But hey, at least I was smart enough to ignore the conventional wisdom. I can't wait for the new ancient samples from the Pontic-Caspian steppe that David Anthony featured in his talks recently. Once I have them we'll be able to work out the details here for ourselves. See also... Ahead of the pack Ancient DNA vs Ex Oriente Lux Understanding the Eneolithic steppe
Sunday, March 14, 2021
A comedy of errors
A couple of years ago, the authors of a paper about a group of Iron Age nomads from the site of Shirenzigou, in the eastern Tian Shan, made a mistake. They wrongly assigned two of these nomads to Y-haplogroup R1b-M269.
This faux pas made them believe that the Shirenzigou nomads were closely related to the M269-rich population associated with the Afanasievo culture.
Indeed, since the Afanasievo culture was often credited with the spread of Tocharian languages to the Tarim Basin, these authors, led by Chao Ning, also concluded that the Shirenzigou nomads were potentially the missing link between the Afanasievo culture and the Tocharians (see here).
Moreover, Ning et al. used formal statistics to argue that the Shirenzegou nomads harbored Afanasievo-related genome-wide ancestry, rather than Sintashta-related genome-wide ancestry, despite the fact that the latter ancestry was widespread in the Tian Shan and surrounds during the Bronze and Iron ages. Soon after, another group of authors, led by Chuan-Chao Wang, also went out of their way to link the Shirenzigou nomads to the Afanasievo people with genome-wide DNA using formal statistics (see here).
Interestingly, one of the Shirenzigou nomads belongs to Y-haplogroup R1a-Z93, which is an obvious Sintashta-related lineage. Both Ning et al. and Wang et al. missed this important fact.
They also missed the key fact that the R1b lineage found in the Shirenzigou nomads actually belongs to an Inner Asian subclade, which is only very distantly related to the originally Eastern European R1b-M269.
Now, formal stats are a very useful tool for studying genome-wide ancestry. But they're not infallible, and that's actually something of an understatement. Indeed, if you don't run sanity checks when using formal stats, you're likely to come to some unusual, even arse about face, conclusions. Uniparental markers, like Y-chromosome haplogroups, can provide a robust sanity check when running formal stats on genome-wide data.
One problem with formal stats is that Sintashta-related ancestry often looks very much like Afanasievo-related ancestry when it's mixed with indigenous Central Asian ancestry. Basically, the reason why this happens is that the Central Asian ancestry dampens the Early European Farmer (EEF) signal in the Sintashta-related ancestry.
This is an artifact that once caused scientists at Harvard to believe that Central Asian Scythians and present-day South Asians lacked Sintashta-related ancestry.
Unfortunately, since the publication of the Ning et al. paper, a consensus has emerged in academia that the Shirenzigou nomads are indeed the missing link between the Afanasievo culture and the Tocharians. But, let's be objective and honest here, it's a consensus based on nothing more than a comedy of errors.
On the other hand, me and most of the commentators at this blog have formed opinions about the Shirenzigou nomads that are totally at odds with the academic consensus, that:
- they're a complex mixture of Sintashta-related, indigenous Central Asian and Tibetan-related ancestries, with no clear, unambiguous signal of Afanasievo-related ancestry - they weren't the speakers of Proto-Tocharian or even related in any specific way to the Tocharians - they were probably the speakers of a now extinct Indo-Iranian language, and, at least based on geographic proximity, possibly related to the Yuezhi.Feel free to make up your own mind. But for me, the question of how Tocharian languages ended up in the Tarim Basin remains wide open. I admit though, I'm currently quite partial to the idea floated here by commentator Copper Axe that the Chemurchek culture may have had something to do with it. See also... Don't believe everything you read in peer reviewed papers
Labels:
Afanasievo,
ancient DNA,
Botai,
Central Asia,
China,
proto-Tocharian,
R1a-Z93,
R1b-M269,
Shirenzigou,
Shirenzigou nomads,
Sintashta,
Sintashta-Petrovka culture,
Tarim Basin,
Tian Shan,
Tocharian
Saturday, February 13, 2021
The Uralic cline with kra001 - no projection this time
A whole lot of nonsense was posted online, often by people who should've known better, after I claimed that kra001 was a solid proxy for a proto-Uralic genome (see here).
For those of you who still don't get it, below are three Principal Component Analysis (PCA) plots featuring Uralic speakers and other present-day Eurasians. Kra001 is also there. These graphs are based on genotype data not reprocessed Global25 data. The relevant datasheet is available here.
Compared to my previous PCA with kra001, here I included a bigger range of East Eurasian populations to help mitigate the effects of extreme genetic drift in some of the Siberian groups, at least on the first few Principal Components (PCs). Moreover, kra001 wasn't projected onto PCs computed with modern-day samples, so he was free to influence the outcome of the PCA.
Note the east to west clines made up largely of Uralic speaking groups on the first two plots. These plots are based on PCs 1/2 and 1 /3, respectively. The third plot, based on PCs 1/4, is more complex and thus more difficult to interpret, but it also manages to isolate many of the Uralic populations from the others.
The Uralic-specific clines do intersect with the clines and clusters formed by the other linguistic groups. However, based on the three plots, the Yeniseian-speaking Kets are the only Asian group that can plausibly be confused for Uralic speakers.
Importantly, apart from the Kets, kra001 is the only Asian individual who shifts his position on all three plots as if he were a Uralic speaker. This might well be a coincidence, and we'll never know what language was spoken by kra001, but it does suggest to me that his genome is a solid proxy for a proto-Uralic genome.
See also...
First taste of Early Medieval DNA from the Ural region
The BOO people: earliest Uralic speakers in the ancient DNA record?
Fresh off the sledge
Friday, February 5, 2021
Finally, a proto-Uralic genome
Obviously, genes don't speak languages, people do. But sometimes it's possible to associate a linguistic group with a very specific genetic signature.
A while ago many of us in the blogosphere spotted an uncanny connection between the Uralic language family, Y-haplogroup N-L1026 and Nganasan-like genome-wide genetic ancestry.
As a result, we expected a Nganasan-like population rich in N-L1026 to eventually appear in the ancient DNA record, probably somewhere in Siberia and in burials from a likely proto-Uralic archeological culture. This hasn't happened yet, but we now have direct evidence that such a population must have existed somewhere deep in Siberia as early as the Bronze Age.
Kra001, whose genome was published recently along with Kilinc et al., belongs to a pre-N-L1026 lineage and, at least in terms of genome-wide genetic structure, could well be from a population directly ancestral to present-day Nganasans. Of course, the Nganasan language is part of the Samoyedic branch of Uralic.
Below is a series of Principal Component Analyses (PCA) featuring kra001. He's labeled RUS_Krasnoyarsk_BA, after the location and age of his burial. Note the obvious Uralic cline running across the plots. That is, from west to east. Kra001 is positioned at the end of this cline very close to a small cluster of Nganasans. To see interactive versions of the plots, paste the Global25 coordinates here into the relevant field here.
Admittedly, there's no way of knowing whether this individual spoke proto-Uralic or not. Indeed, he may have spoken something totally unrelated. The important point is that the very specific genetic signature shared by almost all present-day Uralic speakers, except perhaps Hungarians, is now finally represented in the ancient DNA record. And I can reveal to you that we'll soon be seeing many more ancients very similar to kra001 in upcoming papers.
See also...
The Uralic cline with kra001 - no projection this time
The BOO people: earliest Uralic speakers in the ancient DNA record?
Fresh off the sledge
Wednesday, January 27, 2021
The great shift
Here's a Principal Component Analysis (PCA) featuring some of the ancients from the recent Saag et al. paper at Science Advances. To see an interactive version of the plot paste the Global25 coordinates here into the relevant field here.
Note that the Fatyanovo culture agropastoralists, who are rich in Y-haplogroup R1a and steppe ancestry, cluster with present-day Eastern Europeans. On the other hand, the Volosovo culture singleton sits near the European hunter-gatherer cline that no longer exists.
This Volosovo individual belongs to Y-haplogroup Q1a. However, most of the Volosovo males whose genomes are soon to be published belong to Y-haplogroup R1b.
Thus, in much of Eastern Europe during the Bronze Age, agropastoralists rich in R1a and steppe ancestry replaced hunter-gatherers rich in R1b and with no steppe ancestry. Of course, that's not where the story ends, but I'll get back to that later this year.
By the way, the relatively high coverage Fatyanovo Y-chromosome sequences are being analyzed at YFull. You can check out the results here.
See also...
Labels:
ancient DNA,
Bronze Age,
Eastern Europe,
Fatyanovo,
Fatyanovo-Balanovo culture,
Indo-Aryan,
Indo-European,
Indo-Iranian,
Pontic-Caspian steppe,
Q1a,
R1a,
R1a-Z93,
R1b,
steppe ancestry,
Volosovo culture
Sunday, January 17, 2021
A tantalizing link
A new paper at PLoS ONE reports on the first human genomes reliably associated with the Single Grave culture (SGC). They were sequenced from remains in a burial at Gjerrild, Denmark, roughly dating to 2,500 BCE.
Surprisingly, one of the male genomes belongs to Y-haplogroup R1b-V1636, which is an exceedingly rare marker both in ancient and present-day populations.
However, the results do make sense, because the earliest instances of R1b-V1636 are in three Eneolithic males from burial sites on the Pontic-Caspian (PC) steppe in Eastern Europe, which is precisely where one would expect to find the paternal ancestors of the SGC population. The SGC, of course, is the westernmost variant of the Corded Ware culture (CWC), and there's very little doubt nowadays that the CWC had its roots on the PC steppe.
A Copper Age individual from Arslantepe in central Anatolia also belongs to R1b-V1636, which suggests that Northern Europe shared a very specific link with Anatolia via Eastern Europe during a period generally regarded to have been the time of early Indo-European dispersals.
Numerous SGC barrows or kurgans dot the landscape in what are now the Netherlands, northwestern Germany and Denmark. Unfortunately, most SGC human remains have been eaten up by the acidic soils that exist in this area.
Citation: Egfjord AF-H, Margaryan A, Fischer A, Sjögren K-G, Price TD, Johannsen NN, et al. (2021) Genomic Steppe ancestry in skeletons from the Neolithic Single Grave Culture in Denmark. PLoS ONE 16(1): e0244872. https://doi.org/10.1371/journal.pone.0244872
See also...
Maykop ancestry in Copper Age Arslantepe
That old chestnut: Northeast vs Northwest Euros
In the last comment thread reader Greg put forth this question:
David, when are you going to explain the genetic discrepancy between Northeastern and Northwestern Europeans? You know, the one that people believe is due to Baltic Hunter-Gatherer admixture, whereas you believe it is due to genetic drift? You ought to make a post about this issue at some point, because a lot of people are wondering what's causing the differences.Well, Greg, this issue has been discussed to the proverbial death here and elsewhere. In fact, there were two posts and rather lengthy comment threads on the same topic at this blog just a few months ago. See here and here. Nevertheless, it seems that a fair number of people are still befuddled, so I'm going to try to explain this one last time, as briefly as a I can using just a handful of f4-stats. Admittedly, Northeast Europeans generally do pack higher levels of indigenous European hunter-gatherer ancestry than Northwest Europeans. This is especially true of Balts, who show more of this type of ancestry than even Scandinavians in practically every type of analysis. The f4-stats below back this up unambiguously. Note the significantly positive (>3) Z scores, which suggest that Latvians and Lithuanians harbor more Baltic hunter-gatherer-related ancestry than Norwegians and Swedes.
Chimp Baltic_HG Norwegian Latvian 0.001301 7.114 Chimp Baltic_HG Swedish Latvian 0.001017 4.205 Chimp Baltic_HG Norwegian Lithuanian 0.001023 7.341 Chimp Baltic_HG Swedish Lithuanian 0.000763 3.408Greg, I know what you're thinking: the naysayers are right! But wait, because there's a twist to this tale. Check out these f4-stats:
Chimp Baltic_HG Norwegian Belarusian 0.000265 1.934 Chimp Baltic_HG Swedish Belarusian 0.000152 0.7 Chimp Baltic_HG Norwegian Polish 6.4E-05 0.519 Chimp Baltic_HG Swedish Polish -0.000235 -1.074Please note, Greg, that none of the Z scores reach significance, which means that these Northwest Europeans and Slavs are symmetrically related to Baltic_HG. They're also symmetrically related to other relevant ancient groups such as the Yamnaya steppe herders. This, of course, suggests that they harbor very similar levels of basically the same ancient genetic components.
Chimp Karelia_HG Norwegian Belarusian 0.000136 0.844 Chimp Karelia_HG Swedish Belarusian 7.9E-05 0.32 Chimp Karelia_HG Norwegian Polish -4.7E-05 -0.304 Chimp Karelia_HG Swedish Polish -0.000134 -0.54 Chimp Yamnaya_Samara Norwegian Belarusian -0.000134 -1.085 Chimp Yamnaya_Samara Swedish Belarusian -6.6E-05 -0.34 Chimp Yamnaya_Samara Norwegian Polish -0.000225 -1.995 Chimp Yamnaya_Samara Swedish Polish -0.000311 -1.574 Chimp Barcin_N Norwegian Belarusian -0.000335 -2.809 Chimp Barcin_N Swedish Belarusian -0.000284 -1.491 Chimp Barcin_N Norwegian Polish -0.000222 -2.057 Chimp Barcin_N Swedish Polish -0.000318 -1.662 Chimp Baikal_N Norwegian Belarusian 0.000186 1.3 Chimp Baikal_N Swedish Belarusian -7E-05 -0.33 Chimp Baikal_N Norwegian Polish -4.6E-05 -0.351 Chimp Baikal_N Swedish Polish -0.000477 -2.277Interestingly, pairing up Ukrainians with English samples from Cornwall and Kent produces similar outcomes. But that's because most ancient ancestry proportions in Europe show a closer correlation with latitude than longitude.
Chimp Baltic_HG English_Cornwall Ukrainian 0.000282 2.242 Chimp Baltic_HG English_Kent Ukrainian 0.000225 1.748 Chimp Karelia_HG English_Cornwall Ukrainian 0.000323 2.175 Chimp Karelia_HG English_Kent Ukrainian 0.000239 1.634 Chimp Yamnaya_Samara English_Cornwall Ukrainian -6.6E-05 -0.569 Chimp Yamnaya_Samara English_Kent Ukrainian -0.000112 -0.977 Chimp Barcin_N English_Cornwall Ukrainian -0.000519 -4.641 Chimp Barcin_N English_Kent Ukrainian -0.000598 -5.232 Chimp Baikal_N English_Cornwall Ukrainian 0.000385 2.874 Chimp Baikal_N English_Kent Ukrainian 0.00036 2.836Now, Greg, if at least in terms of genetic ancestry, Latvians, Lithuanians, Belarusians, Poles and Ukrainians all qualify as Northeast Europeans, then what makes them different, as a group, from Northwest Europeans? Do you believe that the key factor is admixture from Baltic hunter-gatherers? Or is it genetic drift? Of course, considering all of the f4-stats above, logic dictates that it must be relatively recent genetic drift. Keep in mind, however, that this only applies to Balto-Slavic speaking Northeast Europeans without significant Uralian ancestry. Overall, Uralic speakers have a more complex population history, and indeed genetic differences between them and Northwest Europeans are in large part due to somewhat different ancestry proportions and also Siberian admixture. See also... So who's the most (indigenous) European of us all?
Thursday, January 14, 2021
David Anthony on Y-haplogroup R1a
Archeologist David Anthony has a new theory which attempts to explain why Y-haplogroup R1a hasn't yet been found in any Yamnaya graves. Basically, he thinks that it was carried by Yamnaya men who weren't buried in kurgans, because they were part of a social underclass, and so their remains are now difficult to locate. See here.
This is an interesting attempt to find a socio-archeological solution to a genetic question, but it's unnecessarily complicated and, in fact, also unnecessary.
The important thing to understand about R1a is that it's rarely seen in the ancient DNA record before the rise of the Corded Ware culture (CWC). Moreover, the vast majority of the R1a lineages in the world today belong to the R1a-M417 subclade, which is a relatively young (Eneolithic era?) marker and closely associated with the CWC population and its rapid expansion.
Indeed, modern R1a lineages show a very strong star-like phylogeny indicative of a series of rapid and massive expansions starting from a handful of lineages only a few thousand years ago.
So if R1a was actually present in the Yamnaya population, then the obvious reason why it hasn't yet been found in any Yamnaya remains is because it was only carried by a very small group of Yamnaya men. Simple as that.
Its expansions from the Pontic-Caspian (PC) steppe, predominantly via the highly successful R1a-M417, may have coincidentally and rather ironically started in a socially disenfranchised Yamnaya clan.
But my view is that R1a-M417 just happened to be present in a small group of early Yamnaya or Yamnaya-related males who came up with an economic package that allowed them to expand out of the PC steppe like no one else before them, and so they did just that.
Anthony is currently collaborating on a new paper about the Eneolithic era on the PC steppe with scientists from Harvard's David Reich Lab (see here). I'm really hoping that they get this right.
See also...
Fatyanovo as part of the wider Corded Ware family
Subscribe to:
Posts (Atom)