R1a-M417 and R1b-L51 are by far the most important Y-chromosome haplogroups in Europe today. More precisely, R1a-M417 dominates in Eastern Europe, while R1b-L51 in Western Europe.
It's been obvious for a while now, at least to me, that both of these Y-haplogroups are closely associated with the men of the Late Neolithic Corded Ware culture (CWC). Indeed, in my mind they're the main genetic signals of its massive expansion, probably from a homeland somewhere north of the Black Sea in what is now Ukraine.
I'm still not exactly sure how the east/west dichotomy between R1a and R1b emerged in Europe, but, thanks to a
new paper by Papac et al. at
Science Advances, at least now I have a working hypothesis about that. Below is a quote from the said paper, emphasis is mine:
In addition to autosomal genetic changes through time, we observe a sharp reduction in Y-chromosomal diversity going from five different lineages in early CW to a dominant (single) lineage in late CW (Fig. 4A). We used forward simulations to explore the demographic scenarios that could account for the observed reduction in Y-chromosomal diversity. Performing 1 million simulations of a population with a starting frequency of R1a-M417(xZ645) centered around the observed starting frequency in Bohemia_CW_Early (3 of 11, 0.27), we assessed the plausibility of this lineage reaching the observed frequency in Bohemia_CW_Late (10 of 11, 0.91) in the time frame of 500 years under a model of a closed population and random mating (Materials and Methods). We reject the “neutral” hypothesis, i.e., that this change in frequency occurred by chance, given a wide range of plausible population sizes. Instead, our results suggest that R1a-M417(xZ645) was subject to a nonrandom increase in frequency, resulting in these males having 15.79% (4.12 to 44.42%) more surviving offspring per generation relative to males of other Y-haplogroups. We also find that this change in Y chromosome frequency is extreme compared to the changes in allele frequencies at fully covered autosomal 1240k sites within the same males, suggesting a process that disproportionately affected Y-chromosomal compared to autosomal genetic diversity, ruling out a population bottleneck as the likely cause. Our results suggest that the Y-lineage diversity in early CW males was supplanted by a nonrandom process [selection, social structure, or influx of nonlocal R1a-M417(xZ645) lineages] that drove the collapse in Y-chromosomal diversity. A simultaneous decline of Y-chromosomal diversity dating to the Neolithic has been observed across most extant Y-haplogroups (64), possibly due to increased conflict between male-mediated patrilines (65). We view that changes in social structure (e.g., an isolated mating network with strictly exclusive social norms) could be an alternative cause but would be difficult to distinguish in the underlying model parameters.
Right, so even though the CWC was clearly a community of closely related groups, there must have been some competition between its different clans. And since these clans were highly patriarchal and patrilineal, this competition probably led to different paternal lineages dominating different parts of the CWC horizon, with M417 becoming especially common in the east and L51 in the west.
Of course, the expansions of post-Corded Ware groups, such as the M417-rich Slavs in Eastern Europe and L51-rich Celts in Western Europe, were also instrumental in creating Europe's R1a/R1b dichotomy, but obviously these groups were in large part the heirs of the CWC.
By the way, most of the samples from Papac et al. are already in the Global25 datasheets linked
here. Look for the labels listed
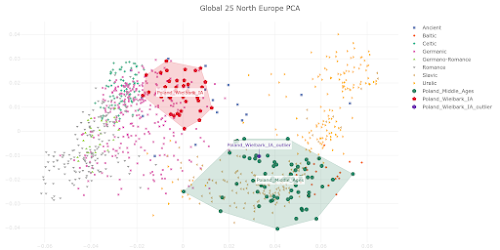
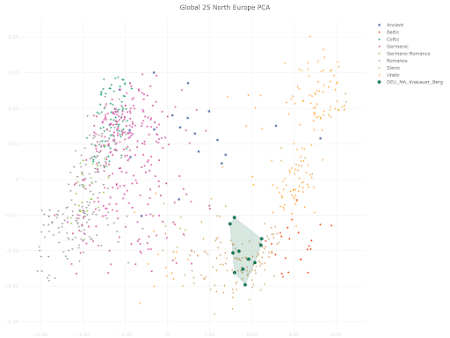
here. Below is a plot made from the Global25 data courtesy of regular commentator Matt.
Citation: L. Papac, M. Ernée, M. Dobeš, M. Langová, A. B. Rohrlach, F. Aron, G. U. Neumann, M. A. Spyrou, N. Rohland, P. Velemínský, M. Kuna, H. Brzobohatá, B. Culleton, D. Daněček, A. Danielisová, M. Dobisíková, J. Hložek, D. J. Kennett, J. Klementová, M. Kostka, P. Krištuf, M. Kuchařík, J. K. Hlavová, P. Limburský, D. Malyková, L. Mattiello, M. Pecinovská, K. Petriščáková, E. Průchová, P. Stránská, L. Smejtek, J. Špaček, R. Šumberová, O. Švejcar, M. Trefný, M. Vávra, J. Kolář, V. Heyd, J. Krause, R. Pinhasi, D. Reich, S. Schiffels, W. Haak,
Dynamic changes in genomic and social structures in third millennium BCE central Europe. Sci. Adv. 7, eabi6941 (2021).
See also...
On the origin of the Corded Ware people
Understanding the Eneolithic steppe
Conan the Barbarian probably belonged to Y-haplogroup R1a