search this blog
Tuesday, May 28, 2019
North African ancestry in a British Bell Beaker?
A new thesis uploaded to the University of Huddersfield Repository suggests that there might be "Near Eastern/North African ancestry in a Bell Beaker individual from northeastern England" (see here).
If true, this wouldn't be a shocking outcome, but certainly an interesting one. Do any of the British Beakers in the Global25 datasheets show this type of ancestry?
See also...
A Bell Beaker superhighway
The Boscombe Bowmen
Migration of the Bell Beakers—but not from Iberia (Olalde et al. 2018)
Friday, May 24, 2019
More on the association between Uralic expansions and Y-haplogroup N
Genes don't speak languages, people do. Thus, associations between genetic markers and languages may often not be easy to discern, especially with time. This is the case when it comes to Y-chromosome haplogroup N and the Hungarian language.
I briefly discussed this problem not long ago in the context of new ancient DNA samples from medieval Hungary (see here). Today, a detailed paper on the topic by Post et al. was published at Scientific Reports (open access here). It brings together evidence from modern and ancient DNA, linguistics and archeology to argue that Hungarian was introduced into the Carpathian Basin during the Middle Ages by migrants from near the Ural Mountains rich in Y-haplogroup N3a4-B539. Below is the paper abstract, emphasis is mine:
Hungarians who live in Central Europe today are one of the westernmost Uralic speakers. Despite of the proposed Volga-Ural/West Siberian roots of the Hungarian language, the present-day Hungarian gene pool is highly similar to that of the surrounding Indo-European speaking populations. However, a limited portion of specific Y-chromosomal lineages from haplogroup N, sometimes associated with the spread of Uralic languages, link modern Hungarians with populations living close to the Ural Mountain range on the border of Europe and Asia. Here we investigate the paternal genetic connection between these spatially separated populations. We reconstruct the phylogeny of N3a4-Z1936 clade by using 33 high-coverage Y-chromosomal sequences and estimate the coalescent times of its sub-clades. We genotype close to 5000 samples from 46 Eurasian populations to show the presence of N3a4-B539 lineages among Hungarians and in the populations from Ural Mountain region, including Ob-Ugric-speakers from West Siberia who are geographically distant but linguistically closest to Hungarians. This sub-clade splits from its sister-branch N3a4-B535, frequent today among Northeast European Uralic speakers, 4000–5000 ya, which is in the time-frame of the proposed divergence of Ugric languages.Post et al., Y-chromosomal connection between Hungarians and geographically distant populations of the Ural Mountain region and West Siberia, Scientific Reports 9, Article number: 7786 (2019), DOI: https://doi.org/10.1038/s41598-019-44272-6 See also... Hungarian Conquerors were rich in Y-haplogroup N (Fóthi et al. 2020) On the association between Uralic expansions and Y-haplogroup N Ancient DNA confirms the link between Y-haplogroup N and Uralic expansions
Labels:
Carpathian Basin,
Central Europe,
Finnic,
Finno-Ugric,
Hungarian,
Hungarian Plain,
Indo-European,
N1c,
N3a,
N3a4-B539,
Northeastern Europe,
Proto-Indo-European,
Proto-Uralic,
Ugric,
Ural Mountains,
Uralic
Sunday, May 19, 2019
Who were the people of the Nordic Bronze Age?
Ancient DNA has revealed that large scale migrations and population replacements have often accompanied major cultural changes in prehistoric Europe. But, for now, my opinion is that the formation of the archeologically ostentatious Nordic Bronze Age wasn't associated with any significant foreign gene flow into Scandinavia. I've tested this as best as I could with the few relevant ancient samples that are currently available.
For instance, below are among the most successful qpAdm mixture models that I was able find for various ancient Scandinavian groups dating back to the local Middle Neolithic (MN) period. The Nordic Bronze Age population is represented by three individuals labeled Nordic_BA. Unfortunately, the guy pictured above, from the famous Borum Eshøj barrow burial in what is now Denmark, didn't make the cut. For more details about my sampling and labeling strategies refer to the text file here.
Nordic_MN_B CWC_CZE 0.822±0.059 POL_Globular_Amphora 0.178±0.059 chisq 14.478 tail prob 0.341086 Full output SWE_Battle_Axe CWC_Baltic_early 0.662±0.028 POL_Globular_Amphora 0.338±0.028 chisq 11.234 tail prob 0.591189 Full output Nordic_LN Nordic_MN_B 0.928±0.069 SWE_TRB 0.072±0.069 chisq 12.139 tail prob 0.516307 Full output Nordic_BA Nordic_LN 0.851±0.061 SWE_TRB 0.149±0.061 chisq 10.897 tail prob 0.619475 Full outputIt's impossible to successfully model the ancestries of Nordic_MN_B and SWE_Battle_Axe simply with the populations that were living in Scandinavia before them. Therefore, it's likely that they were migrants or the recent descendants of migrants to Scandinavia. But there's nothing surprising about that, because they're archeologically associated with the Corded Ware culture (CWC), which has always been seen as intrusive to Scandinavia from the south and east. Conversely, it's easy to produce statistically sound mixture models for both Nordic_LN and Nordic_BA exclusively with earlier Scandinavian populations. Indeed, based on the outgroups or right pops that I'm using, Nordic_LN is almost indistinguishable from Nordic_MN_B, and the same can be said of Nordic_BA in regards to Nordic_LN. Of course, if I mixed and matched reference populations from across prehistoric Europe, I could probably come up with some spectacular statistical fits even without the need for any Scandinavians. Essentially that's because Nordic_LN and Nordic_BA are closely related to many earlier and contemporaneous peoples living all the way from the Atlantic facade to the Ural Mountains. My point, however, is that this isn't crucial, despite the dearth of ancient samples from Scandinavia. This is how things look in a Principal Component Analysis (PCA) of Northern European genetic variation based on my Global25 data. Strikingly, Nordic_MN_B, SWE_Battle_Axe, Nordic_LN and Nordic_BA more or less recapitulate the cluster made up of present-day Swedish samples. The relevant datasheet is available here. Granted, two of the Nordic_BA samples sit just south of the Swedes, no doubt due to their slightly higher ratios of Neolithic farmer (SWE_TRB-related) ancestry, but this is also an area of the plot that many present-day Danes call home (not shown, because I don't have any suitable academic Danish samples to run). I'll eat my hat if it turns out that Scandinavia experienced a major population shift (say, more than a collateral ~10%) during the LN and/or BA periods. And I'll post a clip of it online too. Update 27/08/2019: Four of the samples from the recent Frei et al. paper on human mobility in prehistoric southern Scandinavia are in my Global25 datasheets. So I thought it might be interesting to check whether their strontium isotope ratios correlated with their genomic profiles. In the Principal Component Analysis (PCA) below, RISE61 is a subtle outlier along the horizontal axis compared to the other three Nordic ancients, as well as a Danish individual representative of the present-day Danish gene pool. Also note that RISE61 shows the most unusual strontium isotope ratio (0.712588). The PCA was run with an online tool freely available here. To help drive the point home, here's a figure from Frei et al., edited by me to show the positions of RISE47, RISE61 and RISE71. If RISE276 was also in this graph, he'd be sitting well under the "local" baseline, in roughly the same spot along the vertical axis as RISE47. Interestingly, RISE61 belongs to Y-chromosome haplogroup R1a-M417, while RISE47 and RISE276, who appear to have been locals, both belong to R1b-M269. My guess is that RISE61 was a recent migrant from a more northerly part of Scandinavia dominated by the Battle-Axe culture (BAC). The BAC population was probably rich in R1a-M417 because it moved into Scandinavia from the Pontic-Caspian steppe via the East Baltic. This is what Frei et al. say about RISE61 and his burial site:
The double passage grave of Kyndeløse (Fig 1, S1 File) located on the island of Zealand yielded 70 individuals as well as a large number of grave goods, including flint artefacts, ceramics, and tooth and amber beads. We conducted strontium isotope analyses of seven individuals from Kyndeløse encompassing a period of c. 1000 years, indicating the prolonged use of this passage grave. The oldest of the seven individuals is a female (RISE 65) from whom we measured a “local” strontium isotope signature ( 87 Sr/ 86 Sr = 0.7099). Similar values were measured in five other individuals, including adult males and females. Only a single individual from Kyndeløse, an adult male (RISE 61) yielded a somewhat different strontium isotope signature of 87 Sr/ 86 Sr = 0.7126 which seems to indicate a non-local provenance. The skull of this male individual revealed healed porosities in the eye orbits, cribra orbitalia, a condition which is possibly linked to a vitamin deficiency during childhood, such as iron deficiency.By the way, RISE47 was buried in a flat grave, which suggests that he was a commoner. RISE276 was found in a peat bog in Trundholm, where the famous Trundholm sun chariot was discovered (see here). He may have been a human sacrifice. Citation... Frei KM, Bergerbrant S, Sjögren K-G, Jørkov ML, Lynnerup N, Harvig L, et al. (2019) Mapping human mobility during the third and second millennia BC in present-day Denmark. PLoS ONE 14(8): e0219850. https://doi.org/10.1371/journal.pone.0219850 See also... They came, they saw, and they mixed Children of the Divine Twins The mystery of the Sintashta people
Labels:
ancient DNA,
Battle-Axe culture,
chariot,
Corded Ware Culture,
Denmark,
I1,
Indo-Aryan,
Nordic,
Nordic Bronze Age,
Northern Europe,
R1a-Z284,
R1b-M269,
R1b-U106,
Scandinavia,
Single Grave culture,
Sweden,
TRB
Thursday, May 16, 2019
Fresh off the sledge
As things stand, the closest individual to a Proto-Uralic speaker in the ancient DNA record is arguably 0LS10 from an Iron Age tarand grave in what is now Estonia. I say that because:
- isotopic data suggest that 0LS10 wasn't born where he died, and considering his elevated Siberian ancestry relative to earlier and most contemporaneous Baltic ancients, he was very likely a migrant to the Baltic region from the east - the tarand grave tradition appears to be specifically a Finnic (west Uralic) phenomenon that probably spread from the Volga-Oka region, which is just west of where most people place the Proto-Uralic homeland - 0LS10 belongs to Y-chromosome haplogroup N-L1026, a paternal marker that is especially closely associated with Uralic-speaking populations and probably only appeared in the East Baltic region during the transition from the Bronze Age to the Iron Age
You can find more background info about 0LS10 and other relevant samples in Saag et al. 2019 (see here). This is where he sits in my Principal Component Analyses (PCA) focusing on fine scale Northern European genetic diversity. The relevant datasheets are available here and here, respectively.
Note that 0LS10 doesn't cluster strongly with any ancient or modern populations. To investigate this in more detail I ran a series of two-way qpAdm analyses, testing tens of ancient individuals and populations as potential admixture sources. These two models stood out above the rest in terms of their statistical fits, chronology and overall plausibility.
Baltic_EST_IA_0LS10 Baltic_EST_BA 0.826±0.045 RUS_Sintashta_MLBA_o1 0.174±0.045 chisq 12.527 tail prob 0.564048 Full output Baltic_EST_IA_0LS10 Baltic_EST_BA 0.683±0.102 RUS_Mezhovskaya 0.317±0.102 chisq 13.811 tail prob 0.463864 Full outputPlease note that RUS_Sintashta_MLBA_o1 isn't representative of the Sintashta culture population as a whole. It's a group of the most extreme genetic outliers among the Sintashta samples, and they may or may not have been Uralic speakers (see here). Interestingly, the Mezhovskaya culture population is generally associated with the Ugric branch of the Uralic language family. I was also able to closely replicate these results with the Global25/nMonte method; down to almost one per cent. However, the statistical fits (distances) are poor, probably because the reference populations aren't the real mixture sources. This is in line with the fact that their Y-haplogroups are Q1a, R1a and R1b, rather than any type of N.
Baltic_EST_IA:0LS10 Baltic_EST_BA,83.8 RUS_Sintashta_MLBA_o1,16.2 distance%=4.7955 Baltic_EST_IA:0LS10 Baltic_EST_BA,69.8 RUS_Mezhovskaya,30.2 distance%=3.5783
I do realize that two Bronze Age samples from Bolshoy Oleni Ostrov, Kola Peninsula, belong to N-L1026, but adding them to my mixture models doesn't help. Little wonder, because the Kola Peninsula lies within the Arctic Circle, and I'm pretty sure that 0LS10 and his N-L1026 came from somewhere just north of the mixture cline marked on the map below. Unfortunately, I can't test this directly yet due to the scarcity of ancient samples from this region.
Labels:
ancient DNA,
East Baltic,
Estonia,
Finnic,
Global25,
Iron Age,
Mezhovskaya culture,
N-L1026,
N1c,
N3a,
Proto-Uralic,
Q1a,
qpAdm,
R1a,
R1b,
Sintashta,
tarand grave,
Ural Mountains,
Uralic,
Volga-Oka
Saturday, May 11, 2019
Uralic-specific genome-wide ancestry did make a signifcant impact in the East Baltic
I've started analyzing the ancient genotype data from the recent Saag et al. paper on the expansion of Uralic languages and associated spread of Siberian ancestry into the East Baltic region. The paper is freely available here and the data are here.
I really like the paper, but I don't agree with the authors' claim that the appearance of Y-chromosome haplogroup N in what is now Estonia and surrounds during the Iron Age is "not matched by a clear shift in autosomal profiles". In my opinion it certainly is, and, as one would expect, it's a shift towards a genetic profile typical of western Uralic speakers.
I'd say that the easiest way to find this signal is with a Principal Component Analysis (PCA) focusing on fine scale genetic substructures within Northern Europe, like the one below. The relevant datasheet is available here.
Note that the East Baltic Iron Age samples, all from burial sites in what is now Estonia, appear to be peeling away from their Bronze Age predecessors and overlapping strongly with present-day Estonians, who are Uralic speakers. Indeed, the PCA suggests to me that the formation of the greater part of the present-day Estonian gene pool took place in the East Baltic during the transition from the Bronze Age to the Iron Age. That is, when Uralic languages are generally accepted to have arrived in the region from near the Ural Mountains in the east.
I was also able to closely replicate these outcomes with my Global25 data using the method described here. However, in this effort, present-day Estonians are clearly more western than the Estonian Iron Age samples (EST_IA), which might be due to the presence of low level Germanic ancestry in Estonia dating to the medieval period. The relevant datasheet is available here.
Interestingly, the Estonian Bronze Age samples (EST_BA) come from stone-cist graves which are widely hypothesized to have been introduced to the East Baltic from the Nordic Bronze Age civilization. I even recall reading a paper on the topic which claimed that the remains buried in such graves were those of Proto-Germanic-speaking Scandinavian migrants. Well, I haven't had a chance to study these samples in any great detail yet, but considering that in both of the PCA above they're overlapping strongly with Latvian Bronze Age samples (LVA_BA) and sitting far away from the nearest Scandinavians, I'd say they're probably of local stock from way back.
See also...
It was always going to be this way
On the association between Uralic expansions and Y-haplogroup N
Inferring the linguistic affinity of long dead and non-literate peoples: a multidisciplinary approach
Labels:
ancient DNA,
Baltic Sea,
Balts,
Corded Ware Culture,
East Baltic,
Estonia,
Finland,
Finnic,
Global25,
N-L1026,
N1c,
N3a,
Nordic Bronze Age,
PCA,
R1a,
R1a-M417,
Scandinavia,
tarand grave,
Ural Mountains,
Uralic
Thursday, May 9, 2019
It was always going to be this way
The native peoples of the East Baltic - Estonians, Latvians and Lithuanians - are genetically alike and their paternal gene pools are dominated by the same two Y-chromosome haplogroups: R1a and N3a.
Linguistically, however, Estonians are a world apart from Latvians and Lithuanians. That's because the Estonian language belongs to the Uralic language family, which has an obvious North Eurasian character. On the other hand, Latvian and Lithuanian are both classified as Indo-European languages, along with the vast majority of other European languages.
The Uralic and Indo-European language families may or may not descend from the same ancestral tongue, but even if they do, their relationship is very distant.
So how is it that Estonians came to speak a Uralic language? As far back as I can remember, the basic explanation accepted by most people was that Uralic speech arrived in what is now Estonia and neighboring Finland during the Bronze Age with migrants, or perhaps invaders, rich in N3a from somewhere around the Ural Mountains. Conversely, Latvians and Lithuanians were generally assumed to have retained the Indo-European speech of their R1a-rich forefathers from the Pontic-Caspian steppe, who colonized much of Eastern Europe north of the steppe during the Late Neolithic.
Ancient DNA has now uncannily corroborated these theories (for instance, see Mittnik et al. 2018 and, published today, Saag et al. 2019). All it took was a handful of samples from a few relevant sites. I think that's awesome; I love it when sensible, long-standing hypotheses are validated by cutting edge science.
I'll have a lot more to say about the spread of Uralic languages and Uralian genes to the East Baltic when I get my hands on the genotype data from the new Saag et al. paper. I also have a post coming soon about the Nordic Bronze Age. Stay tuned.
Update 10/05/2019: Uralic-specific genome-wide ancestry did make a signifcant impact in the East Baltic
See also...
Late PIE ground zero now obvious; location of PIE homeland still uncertain, but...
Corded Ware people =/= Proto-Uralics (Tambets et al. 2018)
Inferring the linguistic affinity of long dead and non-literate peoples: a multidisciplinary approach
Labels:
ancient DNA,
Baltic Sea,
Balts,
Corded Ware Culture,
East Baltic,
Estonia,
Fennoscandia,
Finland,
Finnic,
Finns,
Latvia,
Lithuania,
N1c,
N3a,
Nordic Bronze Age,
R1a,
R1a-M417,
Scandinavia,
Ural Mountains,
Uralic
Tuesday, May 7, 2019
The execution
Around 2,800 BCE, in what is now southern Poland, a family group of fifteen individuals associated with the Globular Amphora culture (GAC) were massacred. They were probably captured and executed, because each victim was killed with a blow to the head from the same type of weapon, possibly a stone axe, and lacked defensive wounds. The dead were mostly women and children. They were buried in a mass grave, but with great care and very likely by someone who knew them well.
This Late Neolithic mass grave is the focus of a new ancient DNA and archeological research paper at PNAS by Schroeder et al. (see here). The authors tentatively attribute the massacre to the Corded Ware culture (CWC) people, who were expanding rapidly at the time across much of Europe from their homeland on the Pontic-Caspian steppe.
The CWC people may or may not have been responsible; we'll never know for sure. The perpetrators could just as easily have been a competing GAC family group.
In any case, it's interesting to see that the GAC males belong to Y-chromosome haplogroup I2a-L801. This is today a rather uncommon subclade of I2, and almost exclusively found in Germanic-speaking populations, especially Scandinavians. To me this suggests that some Polish GAC males were incorporated into Indo-European-speaking CWC populations that ended up in Scandinavia, and their paternal lineages eventually became a part of the Proto-Germanic gene pool. Admittedly, though, that's just one of many possible scenarios.
See also...
Late PIE ground zero now obvious; location of PIE homeland still uncertain, but...
Corded Ware people =/= Proto-Uralics (Tambets et al. 2018)
Inferring the linguistic affinity of long dead and non-literate peoples: a multidisciplinary approach
Labels:
ancient DNA,
Corded Ware Culture,
execution,
GAC,
Germanic,
Globular Amphora,
I2a,
Indo-European,
Late Neolithic,
mass burial,
mass grave,
massacre,
Poland,
Pontic-Caspian steppe,
R1a-M417,
R1a-Z645,
stone axe
Sunday, May 5, 2019
Conan the Barbarian probably belonged to Y-haplogroup R1a
A fresh batch of Iron Age genomes from across the Eurasian steppes is about to be published along with a new paper at Current Biology. The manuscript, titled Shifts in the Genetic Landscape of the Western Eurasian Steppe Associated with the Beginning and End of the Scythian Dominance, is still under review but freely available here.
Most of the male ancients, including two Cimmerians from the North Pontic steppe, in what is now Ukraine, belong to Y-chromosome haplogroup R1a. Wasn't Conan the Barbarian supposed to be a Cimmerian? From the preprint, emphasis is mine:
The Early Iron Age nomadic Scythians have been described as a confederation of tribes of different origins, based on ancient DNA evidence [1-3]. It is still unclear how much of the Scythian dominance in the Eurasian Steppe was due to movements of people and how much reflected cultural diffusion and elite dominance. We present new whole-genome sequences of 31 ancient Western and Eastern Steppe individuals including Scythians as well as samples pre- and postdating them, allowing us to set the Scythians in a temporal context (in the Western/Ponto-Caspian Steppe). We detect an increase of eastern (Altaian) affinity along with a decrease in Eastern Hunter-Gatherer (EHG) ancestry in the Early Iron Age Ponto- Caspian gene pool at the start of the Scythian dominance. On the other hand, samples of the Chernyakhiv culture postdating the Scythians in Ukraine have a significantly higher proportion of Near Eastern ancestry than other samples of this study. Our results agree with the Gothic source of the Chernyakhiv culture and support the hypothesis that the Scythian dominance did involve a demic component. ... Out of the 31 samples of this study, 16 are male, and with sufficient Y-chromosome coverage for haplogroup assignment (Table S2). R1a (43%) and I (27%) are the two most frequent Y- chromosome hgs in present-day Ukrainians [142]. R1a is also the predominant lineage among Cimmerians, Scy_Ukr and ScySar_SU in our data, and present among Scy_Kaz as well. Thus, although acknowledging our small sample size, the individuals sampled from archaeological context associated with Scythian identity do not appear to stand out from the context of other groups living in the region before and after them. One notable difference from the present is the absence of hg N, nowadays widespread in the Volga-Uralic region and West Siberia as well as among Mongols and Altaians [165-167]; however, this result is consistent with the absence of hg N among Bronze Age and Eneolithic males from the Steppe [168]. In context of their claimed Altaian homeland it is interesting to note that one Scy_Ukr and the single Sar_Cau sample belong to the Q1c-L332 lineage which is a sub-clade of hg Q1c-L330 that today has peak frequency of 68% in Western Mongolians [169] and occurs at 17% in South Altaians [170] while being very rare (<1%) in East European populations and absent elsewhere (https://www.yfull.com/tree/Q-L330/).Järve et al., Shifts in the Genetic Landscape of the Western Eurasian Steppe Associated with the Beginning and End of the Scythian Dominance, Current Biology (preprint), Posted: 6 Mar 2019, http://dx.doi.org/10.2139/ssrn.3346985 Update 12/07/2019: The paper has just been published and is freely available at Current Biology [LINK]. See also... The mystery of the Sintashta people On the association between Uralic expansions and Y-haplogroup N Late PIE ground zero now obvious; location of PIE homeland still uncertain, but...
Labels:
ancient DNA,
Barbarian,
Chernyakhiv culture,
Chernyakhov culture,
Cimmerian,
Eurasian steppe,
Germanic,
Gothic,
Indo-European,
Indo-Iranian,
Iron Age,
North Pontic,
R1a-M417,
R1a-Z645,
Sarmatian,
Scythian,
Thracian
Friday, May 3, 2019
Inferring the linguistic affinity of long dead and non-literate peoples: a multidisciplinary approach
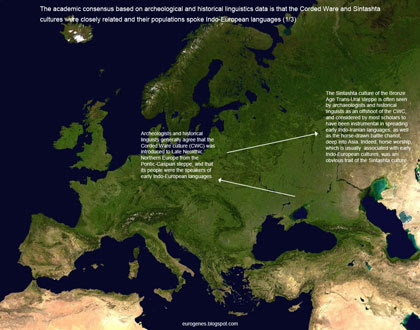
Ancient DNA has treated us to many surprises in recent years. But it has also uncannily corroborated some well established hypotheses that were formulated decades ago from historical linguistics and archeological data. One such hypothesis is that the population associated with the Late Neolithic Corded Ware culture (CWC), and its myriad offshoots, spoke early Indo-European languages and spread them across much of Europe and into the Indian subcontinent.
Below is a series of figures in which I explain why the CWC and its likely close relative, the Sintashta culture, are widely regarded as early Indo-European-speaking cultures, even though their languages aren't attested. To view the images at their maximum size, right click on the thumbs and choose "open link in a new tab".
It's a damn shame that we still don't know where the modern domesticated horse lineage ultimately came from. I'm pretty sure that it came from the Pontic-Caspian steppe, but I was hoping this would be confirmed in the latest paper on horse genomics published today at Current Biology: Tracking Five Millennia of Horse Management with Extensive Ancient Genome Time Series. Nope, the topic wasn't even covered, and no wonder, because the sampling strategy in the paper didn't allow it to be. What we desperately need are samples associated with such archeological cultures as Khvalynsk, Repin, Sredny Stog and Yamnaya. Maybe next time, eh?
See also...
Labels:
ancient DNA,
Arabian horse,
Battle-Axe,
Botai,
chariot,
Corded Ware Culture,
Iberian horse,
Indo-Aryan,
Indo-European,
Iranian,
Northern Europe,
Pontic-Caspian steppe,
R1a-M417,
R1a-Z93,
Sintashta,
Turkoman horse
Subscribe to:
Posts (Atom)