search this blog
Showing posts with label Finnic. Show all posts
Showing posts with label Finnic. Show all posts
Saturday, February 13, 2021
The Uralic cline with kra001 - no projection this time
A whole lot of nonsense was posted online, often by people who should've known better, after I claimed that kra001 was a solid proxy for a proto-Uralic genome (see here).
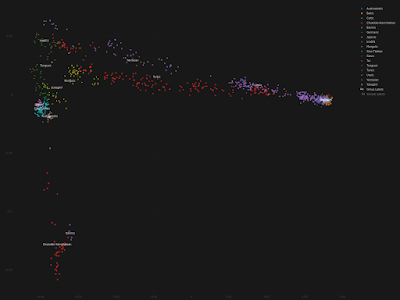
For those of you who still don't get it, below are three Principal Component Analysis (PCA) plots featuring Uralic speakers and other present-day Eurasians. Kra001 is also there. These graphs are based on genotype data not reprocessed Global25 data. The relevant datasheet is available here.
Compared to my previous PCA with kra001, here I included a bigger range of East Eurasian populations to help mitigate the effects of extreme genetic drift in some of the Siberian groups, at least on the first few Principal Components (PCs). Moreover, kra001 wasn't projected onto PCs computed with modern-day samples, so he was free to influence the outcome of the PCA.
Note the east to west clines made up largely of Uralic speaking groups on the first two plots. These plots are based on PCs 1/2 and 1 /3, respectively. The third plot, based on PCs 1/4, is more complex and thus more difficult to interpret, but it also manages to isolate many of the Uralic populations from the others.
The Uralic-specific clines do intersect with the clines and clusters formed by the other linguistic groups. However, based on the three plots, the Yeniseian-speaking Kets are the only Asian group that can plausibly be confused for Uralic speakers.
Importantly, apart from the Kets, kra001 is the only Asian individual who shifts his position on all three plots as if he were a Uralic speaker. This might well be a coincidence, and we'll never know what language was spoken by kra001, but it does suggest to me that his genome is a solid proxy for a proto-Uralic genome.
See also...
First taste of Early Medieval DNA from the Ural region
The BOO people: earliest Uralic speakers in the ancient DNA record?
Fresh off the sledge
Friday, February 5, 2021
Finally, a proto-Uralic genome
Obviously, genes don't speak languages, people do. But sometimes it's possible to associate a linguistic group with a very specific genetic signature.
A while ago many of us in the blogosphere spotted an uncanny connection between the Uralic language family, Y-haplogroup N-L1026 and Nganasan-like genome-wide genetic ancestry.
As a result, we expected a Nganasan-like population rich in N-L1026 to eventually appear in the ancient DNA record, probably somewhere in Siberia and in burials from a likely proto-Uralic archeological culture. This hasn't happened yet, but we now have direct evidence that such a population must have existed somewhere deep in Siberia as early as the Bronze Age.
Kra001, whose genome was published recently along with Kilinc et al., belongs to a pre-N-L1026 lineage and, at least in terms of genome-wide genetic structure, could well be from a population directly ancestral to present-day Nganasans. Of course, the Nganasan language is part of the Samoyedic branch of Uralic.
Below is a series of Principal Component Analyses (PCA) featuring kra001. He's labeled RUS_Krasnoyarsk_BA, after the location and age of his burial. Note the obvious Uralic cline running across the plots. That is, from west to east. Kra001 is positioned at the end of this cline very close to a small cluster of Nganasans. To see interactive versions of the plots, paste the Global25 coordinates here into the relevant field here.
Admittedly, there's no way of knowing whether this individual spoke proto-Uralic or not. Indeed, he may have spoken something totally unrelated. The important point is that the very specific genetic signature shared by almost all present-day Uralic speakers, except perhaps Hungarians, is now finally represented in the ancient DNA record. And I can reveal to you that we'll soon be seeing many more ancients very similar to kra001 in upcoming papers.
See also...
The Uralic cline with kra001 - no projection this time
The BOO people: earliest Uralic speakers in the ancient DNA record?
Fresh off the sledge
Tuesday, January 14, 2020
Hungarian Conquerors were rich in Y-haplogroup N (Fóthi et al. 2020)
Open access at Archaeological and Anthropological Sciences at this LINK. Below is the paper abstract. Emphasis is mine:
According to historical sources, ancient Hungarians were made up of seven allied tribes and the fragmented tribes that split off from the Khazars, and they arrived from the Eastern European steppes to conquer the Carpathian Basin at the end of the ninth century AD. Differentiating between the tribes is not possible based on archaeology or history, because the Hungarian Conqueror artifacts show uniformity in attire, weaponry, and warcraft. We used Y-STR and SNP analyses on male Hungarian Conqueror remains to determine the genetic source, composition of tribes, and kin of ancient Hungarians. The 19 male individuals paternally belong to 16 independent haplotypes and 7 haplogroups (C2, G2a, I2, J1, N3a, R1a, and R1b). The presence of the N3a haplogroup is interesting because it rarely appears among modern Hungarians (unlike in other Finno-Ugric-speaking peoples) but was found in 37.5% of the Hungarian Conquerors. This suggests that a part of the ancient Hungarians was of Ugric descent and that a significant portion spoke Hungarian. We compared our results with public databases and discovered that the Hungarian Conquerors originated from three distant territories of the Eurasian steppes, where different ethnicities joined them: Lake Baikal-Altai Mountains (Huns/Turkic peoples), Western Siberia-Southern Urals (Finno-Ugric peoples), and the Black Sea-Northern Caucasus (Caucasian and Eastern European peoples). As such, the ancient Hungarians conquered their homeland as an alliance of tribes, and they were the genetic relatives of Asiatic Huns, Finno-Ugric peoples, Caucasian peoples, and Slavs from the Eastern European steppes.Fóthi, E., Gonzalez, A., Fehér, T. et al., Genetic analysis of male Hungarian Conquerors: European and Asian paternal lineages of the conquering Hungarian tribes, Archaeol Anthropol Sci (2020) 12: 31. https://doi.org/10.1007/s12520-019-00996-0 See also... On the association between Uralic expansions and Y-haplogroup N More on the association between Uralic expansions and Y-haplogroup N Ancient DNA confirms the link between Y-haplogroup N and Uralic expansions
Labels:
Carpathian Basin,
Central Europe,
Finnic,
Finno-Ugric,
Hungarian,
Hungarian Plain,
Indo-European,
N1c,
N3a,
N3a4-B539,
Northeastern Europe,
Proto-Indo-European,
Proto-Uralic,
Ugric,
Ural Mountains,
Uralic
Sunday, December 1, 2019
Big deal of 2019: ancient DNA confirms the link between Y-haplogroup N and Uralic expansions
The academic consensus is that Indo-European languages first spread into the Baltic region from the Eastern European steppes along with the Corded Ware culture (CWC) and its people during the Late Neolithic, well before the expansion of Uralic speakers into Fennoscandia and surrounds, probably from somewhere around the Ural Mountains.
On the other hand, the views that the Uralic language family is native to Northern Europe and/or closely associated with the CWC are fringe theories usually espoused by people not familiar with the topic or, unfortunately it has to be said, mentally unstable trolls.
The likely close relationship between the CWC expansion and the early spread of Indo-European languages was discussed in several papers in recent years (for instance, see here). This year, we saw the first ancient DNA paper focusing on the transition from the Bronze Age to the Iron Age in the East Baltic, including the likely first arrival of Uralic speech in what is now Estonia.
Published in Current Biology courtesy of Saag et al., the paper showed that the genetic structure of present-day East Baltic populations largely formed in the Iron Age (see here). It was during this time, the authors revealed, that the region experienced a sudden influx of Y-chromosome haplogroup N, which is today common in many Uralic speaking populations and often referred to as a Proto-Uralic marker. Little wonder then that Saag et al. linked this genetic shift in the East Baltic to the westward migrations of early Uralic speakers.
The table below, based on data from the Saag et al. paper, surely doesn't leave much to the imagination about what happened.
Unfortunately, I have to say that the genome-wide analysis in the paper was less informative than it could have been. The authors focused their attention on rather broad genetic components, and, as a result, missed an interesting fine scale distinction between their Bronze Age and Iron Age samples. The spatial maps below, based on my Global25 data for most of the ancients from Saag et al., show what I mean. The hotter the color the higher the genetic similarity between them and present-day West Eurasian populations.
Note that the Bronze Age (Baltic_EST_BA) samples are most similar to the Baltic-speaking, and thus also Indo-European-speaking, Latvians and Lithuanians, rather than the Uralic-speaking Estonians, even though they're from burial sites in Estonia. On the other hand, the Iron Age (Baltic_EST_IA) samples show strong similarity to a wider range of populations, including Estonians and many other Uralic-speaking groups.
See also...
It was always going to be this way
Fresh off the sledge
More on the association between Uralic expansions and Y-haplogroup N
Labels:
ancient DNA,
Corded Ware Culture,
East Baltic,
Estonia,
Fennoscandia,
Finnic,
Finno-Ugric,
Indo-European,
N1c,
N1c1a1a,
N3a,
Northeastern Europe,
Proto-Uralic,
R1a,
R1a-M417,
Siberia,
Uralic,
Urals
Friday, May 24, 2019
More on the association between Uralic expansions and Y-haplogroup N
Genes don't speak languages, people do. Thus, associations between genetic markers and languages may often not be easy to discern, especially with time. This is the case when it comes to Y-chromosome haplogroup N and the Hungarian language.
I briefly discussed this problem not long ago in the context of new ancient DNA samples from medieval Hungary (see here). Today, a detailed paper on the topic by Post et al. was published at Scientific Reports (open access here). It brings together evidence from modern and ancient DNA, linguistics and archeology to argue that Hungarian was introduced into the Carpathian Basin during the Middle Ages by migrants from near the Ural Mountains rich in Y-haplogroup N3a4-B539. Below is the paper abstract, emphasis is mine:
Hungarians who live in Central Europe today are one of the westernmost Uralic speakers. Despite of the proposed Volga-Ural/West Siberian roots of the Hungarian language, the present-day Hungarian gene pool is highly similar to that of the surrounding Indo-European speaking populations. However, a limited portion of specific Y-chromosomal lineages from haplogroup N, sometimes associated with the spread of Uralic languages, link modern Hungarians with populations living close to the Ural Mountain range on the border of Europe and Asia. Here we investigate the paternal genetic connection between these spatially separated populations. We reconstruct the phylogeny of N3a4-Z1936 clade by using 33 high-coverage Y-chromosomal sequences and estimate the coalescent times of its sub-clades. We genotype close to 5000 samples from 46 Eurasian populations to show the presence of N3a4-B539 lineages among Hungarians and in the populations from Ural Mountain region, including Ob-Ugric-speakers from West Siberia who are geographically distant but linguistically closest to Hungarians. This sub-clade splits from its sister-branch N3a4-B535, frequent today among Northeast European Uralic speakers, 4000–5000 ya, which is in the time-frame of the proposed divergence of Ugric languages.Post et al., Y-chromosomal connection between Hungarians and geographically distant populations of the Ural Mountain region and West Siberia, Scientific Reports 9, Article number: 7786 (2019), DOI: https://doi.org/10.1038/s41598-019-44272-6 See also... Hungarian Conquerors were rich in Y-haplogroup N (Fóthi et al. 2020) On the association between Uralic expansions and Y-haplogroup N Ancient DNA confirms the link between Y-haplogroup N and Uralic expansions
Labels:
Carpathian Basin,
Central Europe,
Finnic,
Finno-Ugric,
Hungarian,
Hungarian Plain,
Indo-European,
N1c,
N3a,
N3a4-B539,
Northeastern Europe,
Proto-Indo-European,
Proto-Uralic,
Ugric,
Ural Mountains,
Uralic
Thursday, May 16, 2019
Fresh off the sledge
As things stand, the closest individual to a Proto-Uralic speaker in the ancient DNA record is arguably 0LS10 from an Iron Age tarand grave in what is now Estonia. I say that because:
- isotopic data suggest that 0LS10 wasn't born where he died, and considering his elevated Siberian ancestry relative to earlier and most contemporaneous Baltic ancients, he was very likely a migrant to the Baltic region from the east - the tarand grave tradition appears to be specifically a Finnic (west Uralic) phenomenon that probably spread from the Volga-Oka region, which is just west of where most people place the Proto-Uralic homeland - 0LS10 belongs to Y-chromosome haplogroup N-L1026, a paternal marker that is especially closely associated with Uralic-speaking populations and probably only appeared in the East Baltic region during the transition from the Bronze Age to the Iron Age
You can find more background info about 0LS10 and other relevant samples in Saag et al. 2019 (see here). This is where he sits in my Principal Component Analyses (PCA) focusing on fine scale Northern European genetic diversity. The relevant datasheets are available here and here, respectively.
Note that 0LS10 doesn't cluster strongly with any ancient or modern populations. To investigate this in more detail I ran a series of two-way qpAdm analyses, testing tens of ancient individuals and populations as potential admixture sources. These two models stood out above the rest in terms of their statistical fits, chronology and overall plausibility.
Baltic_EST_IA_0LS10 Baltic_EST_BA 0.826±0.045 RUS_Sintashta_MLBA_o1 0.174±0.045 chisq 12.527 tail prob 0.564048 Full output Baltic_EST_IA_0LS10 Baltic_EST_BA 0.683±0.102 RUS_Mezhovskaya 0.317±0.102 chisq 13.811 tail prob 0.463864 Full outputPlease note that RUS_Sintashta_MLBA_o1 isn't representative of the Sintashta culture population as a whole. It's a group of the most extreme genetic outliers among the Sintashta samples, and they may or may not have been Uralic speakers (see here). Interestingly, the Mezhovskaya culture population is generally associated with the Ugric branch of the Uralic language family. I was also able to closely replicate these results with the Global25/nMonte method; down to almost one per cent. However, the statistical fits (distances) are poor, probably because the reference populations aren't the real mixture sources. This is in line with the fact that their Y-haplogroups are Q1a, R1a and R1b, rather than any type of N.
Baltic_EST_IA:0LS10 Baltic_EST_BA,83.8 RUS_Sintashta_MLBA_o1,16.2 distance%=4.7955 Baltic_EST_IA:0LS10 Baltic_EST_BA,69.8 RUS_Mezhovskaya,30.2 distance%=3.5783
I do realize that two Bronze Age samples from Bolshoy Oleni Ostrov, Kola Peninsula, belong to N-L1026, but adding them to my mixture models doesn't help. Little wonder, because the Kola Peninsula lies within the Arctic Circle, and I'm pretty sure that 0LS10 and his N-L1026 came from somewhere just north of the mixture cline marked on the map below. Unfortunately, I can't test this directly yet due to the scarcity of ancient samples from this region.
Labels:
ancient DNA,
East Baltic,
Estonia,
Finnic,
Global25,
Iron Age,
Mezhovskaya culture,
N-L1026,
N1c,
N3a,
Proto-Uralic,
Q1a,
qpAdm,
R1a,
R1b,
Sintashta,
tarand grave,
Ural Mountains,
Uralic,
Volga-Oka
Saturday, May 11, 2019
Uralic-specific genome-wide ancestry did make a signifcant impact in the East Baltic
I've started analyzing the ancient genotype data from the recent Saag et al. paper on the expansion of Uralic languages and associated spread of Siberian ancestry into the East Baltic region. The paper is freely available here and the data are here.
I really like the paper, but I don't agree with the authors' claim that the appearance of Y-chromosome haplogroup N in what is now Estonia and surrounds during the Iron Age is "not matched by a clear shift in autosomal profiles". In my opinion it certainly is, and, as one would expect, it's a shift towards a genetic profile typical of western Uralic speakers.
I'd say that the easiest way to find this signal is with a Principal Component Analysis (PCA) focusing on fine scale genetic substructures within Northern Europe, like the one below. The relevant datasheet is available here.
Note that the East Baltic Iron Age samples, all from burial sites in what is now Estonia, appear to be peeling away from their Bronze Age predecessors and overlapping strongly with present-day Estonians, who are Uralic speakers. Indeed, the PCA suggests to me that the formation of the greater part of the present-day Estonian gene pool took place in the East Baltic during the transition from the Bronze Age to the Iron Age. That is, when Uralic languages are generally accepted to have arrived in the region from near the Ural Mountains in the east.
I was also able to closely replicate these outcomes with my Global25 data using the method described here. However, in this effort, present-day Estonians are clearly more western than the Estonian Iron Age samples (EST_IA), which might be due to the presence of low level Germanic ancestry in Estonia dating to the medieval period. The relevant datasheet is available here.
Interestingly, the Estonian Bronze Age samples (EST_BA) come from stone-cist graves which are widely hypothesized to have been introduced to the East Baltic from the Nordic Bronze Age civilization. I even recall reading a paper on the topic which claimed that the remains buried in such graves were those of Proto-Germanic-speaking Scandinavian migrants. Well, I haven't had a chance to study these samples in any great detail yet, but considering that in both of the PCA above they're overlapping strongly with Latvian Bronze Age samples (LVA_BA) and sitting far away from the nearest Scandinavians, I'd say they're probably of local stock from way back.
See also...
It was always going to be this way
On the association between Uralic expansions and Y-haplogroup N
Inferring the linguistic affinity of long dead and non-literate peoples: a multidisciplinary approach
Labels:
ancient DNA,
Baltic Sea,
Balts,
Corded Ware Culture,
East Baltic,
Estonia,
Finland,
Finnic,
Global25,
N-L1026,
N1c,
N3a,
Nordic Bronze Age,
PCA,
R1a,
R1a-M417,
Scandinavia,
tarand grave,
Ural Mountains,
Uralic
Thursday, May 9, 2019
It was always going to be this way
The native peoples of the East Baltic - Estonians, Latvians and Lithuanians - are genetically alike and their paternal gene pools are dominated by the same two Y-chromosome haplogroups: R1a and N3a.
Linguistically, however, Estonians are a world apart from Latvians and Lithuanians. That's because the Estonian language belongs to the Uralic language family, which has an obvious North Eurasian character. On the other hand, Latvian and Lithuanian are both classified as Indo-European languages, along with the vast majority of other European languages.
The Uralic and Indo-European language families may or may not descend from the same ancestral tongue, but even if they do, their relationship is very distant.
So how is it that Estonians came to speak a Uralic language? As far back as I can remember, the basic explanation accepted by most people was that Uralic speech arrived in what is now Estonia and neighboring Finland during the Bronze Age with migrants, or perhaps invaders, rich in N3a from somewhere around the Ural Mountains. Conversely, Latvians and Lithuanians were generally assumed to have retained the Indo-European speech of their R1a-rich forefathers from the Pontic-Caspian steppe, who colonized much of Eastern Europe north of the steppe during the Late Neolithic.
Ancient DNA has now uncannily corroborated these theories (for instance, see Mittnik et al. 2018 and, published today, Saag et al. 2019). All it took was a handful of samples from a few relevant sites. I think that's awesome; I love it when sensible, long-standing hypotheses are validated by cutting edge science.
I'll have a lot more to say about the spread of Uralic languages and Uralian genes to the East Baltic when I get my hands on the genotype data from the new Saag et al. paper. I also have a post coming soon about the Nordic Bronze Age. Stay tuned.
Update 10/05/2019: Uralic-specific genome-wide ancestry did make a signifcant impact in the East Baltic
See also...
Late PIE ground zero now obvious; location of PIE homeland still uncertain, but...
Corded Ware people =/= Proto-Uralics (Tambets et al. 2018)
Inferring the linguistic affinity of long dead and non-literate peoples: a multidisciplinary approach
Labels:
ancient DNA,
Baltic Sea,
Balts,
Corded Ware Culture,
East Baltic,
Estonia,
Fennoscandia,
Finland,
Finnic,
Finns,
Latvia,
Lithuania,
N1c,
N3a,
Nordic Bronze Age,
R1a,
R1a-M417,
Scandinavia,
Ural Mountains,
Uralic
Monday, December 3, 2018
On the trail of the Proto-Uralic speakers (work in progress)
Historical linguists have long posited that Fennoscandia was a busy contact zone between early Germanic and Uralic languages. The first ancient DNA samples from what is now Finland have corroborated their inferences, by showing that during the Iron Age the western part of the country was inhabited by a genetically heterogeneous population closely related to both the Uralic-speaking Saami and Germanic-speaking southern Scandinavians.
The samples were sequenced and analyzed by two different teams of researches, and their findings published recently in Lamnidis et al. and Sikora et al. (see here and here, respectively).
This is how most of these ancients, whose remains were excavated from the Levanluhta burial site dated to 300–800 CE, behave in a Principal Component Analysis (PCA) based on my Global25 data. Levanluhta_IA are the Saami-related samples, while Levanluhta_IA_o is an Scandinavian-like outlier. Baltic_IA is an Iron Age individual from what is now Lithuania from the recent Damgaard et al. paper (see here). Note the accuracy of the Global25 data in pinpointing their genetic affinities and also the trajectory of the Levanluhta_IA cluster, which seems to be "pulling" towards Levanluhta_IA_o.
The Saami and Levanluhta_IA are clear outliers from the main Northern European cluster. There are two reasons for this: excess East Asian/Siberian-related ancestry and Saami-specific genetic drift. However, this eastern admixture and genetic drift are shared in varying degrees by other North European populations, especially those that also speak Uralic languages, and this is why they appear to be "pulling" towards the Saami/Levanluhta_IA clusters in my PCA. Thus, what this suggests is that the expansion of Uralic languages across Northeastern Europe was intimately linked with the spread of Siberian-related ancestry into the region.
This idea has been around for a long time and is now becoming even more widely accepted (see here). However, Lamnidis et al. also featured samples from a likely pre-Uralic (1523±87 calBCE) burial site at Bolshoy Oleni Ostrov in the Kola Peninsula, present-day northern Russia, and, perhaps surprisingly, found that they showed even more Siberian-related ancestry than Levanluhta_IA. So what's going on?
I'm confident that this discrepancy can be explained by multiple waves of migrations from the east into Northeastern Europe, possibly before, during and after the time of the people buried at Bolshoy Oleni Ostrov, by pre-Uralic, para-Uralic and/or Proto-Uralic-speaking populations.
Consider the following qpAdm output, in which Levanluhta_IA is just barely modeled successfully as a two-way mixture between Levanluhta_IA_o and Bolshoy_Oleni_Ostrov. The statistical fit improves significantly with the addition of Glazkovo_EBA as a third mixture source. This is an ancient population from near Lake Baikal dated to 4597-3726 BC from the aforementioned Damgaard et al. paper.
Levanluhta_IA Bolshoy_Oleni_Ostrov 0.468±0.036 Levanluhta_IA_o 0.532±0.036 chisq 19.129 tail prob 0.0854706 Full output Levanluhta_IA Bolshoy_Oleni_Ostrov 0.241±0.092 Glazkovo_EBA 0.162±0.059 Levanluhta_IA_o 0.597±0.046 chisq 7.756 tail prob 0.734966 Full outputFor the sake of being complete, I also tested whether Levanluhta_IA_o could be substituted by other similar ancient samples from the neighborhood, including those associated with the Battle-Axe and Corded Ware cultures. There's not much to report; qpAdm returned poor statistical fits and/or implausible ancestry proportions (for the full output from my runs, see here). Baltic_IA did produce a statistically sound model, but with excess Glazkovo_EBA-related ancestry. I also had to drop Bolshoy_Oleni_Ostrov from the analysis to make things work, which suggests to me that the result shouldn't be taken too literally.
Levanluhta_IA Baltic_IA 0.677±0.034 Glazkovo_EBA 0.323±0.034 chisq 8.547 tail prob 0.741095 Full outputSo as far as I can see, the western ancestry in Levanluhta_IA is likely to be mostly of Germanic origin, and thus Indo-European, meaning that it's logical to look east, perhaps far to the east, for the source of its Uralic ancestry. This might seem like a complicated and uncertain task, considering that Levanluhta_IA could well be at least a thousand years younger than the first entry of Uralic speakers into Fennoscandia. However, take a look what happens when I substitute Glazkovo_EBA with a variety of Uralic-speaking populations from around the Ural Mountains, which is where the Proto-Uralic homeland is generally considered to have been located.
Levanluhta_IA Bolshoy_Oleni_Ostrov 0.210±0.091 Khanty 0.283±0.090 Levanluhta_IA_o 0.507±0.035 chisq 7.007 tail prob 0.798532 Full output Levanluhta_IA Bolshoy_Oleni_Ostrov 0.193±0.098 Levanluhta_IA_o 0.495±0.035 Mansi 0.312±0.100 chisq 7.884 tail prob 0.7237 Full output Levanluhta_IA Bolshoy_Oleni_Ostrov 0.300±0.065 Levanluhta_IA_o 0.337±0.072 Mari 0.363±0.121 chisq 8.393 tail prob 0.677705 Full output Levanluhta_IA Bolshoy_Oleni_Ostrov 0.238±0.084 Levanluhta_IA_o 0.553±0.036 Nenets 0.209±0.067 chisq 7.210 tail prob 0.78181 Full output Levanluhta_IA Bolshoy_Oleni_Ostrov 0.302±0.069 Levanluhta_IA_o 0.324±0.081 Udmurt 0.373±0.135 chisq 9.195 tail prob 0.60393 Full outputAll of these models look great, and easily rival the best model with Glazkovo_EBA. Moreover, they make good sense in terms of linguistics. The only problem is that they're anachronistic, because the Uralic-speaking reference populations are younger than Levanluhta_IA. So I can't be certain that they reflect reality without corroboration from ancient DNA. It might turn out, for instance, that a Glazkovo_EBA-like population was already present somewhere deep in Europe before or during the time of Bolshoy_Oleni_Ostrov, while no such population existed around the Ural Mountains until the time of Levanluhta_IA. By the way, it might be important to note that the present-day Finnish samples in my dataset can't be modeled as a mixture between Levanluhta_IA and Levanluhta_IA_o. But they can be modeled as a mixture between Baltic_IA and Levanluhta_IA. I don't know which part of Finland they're from exactly; probably all over the place, so it'd be useful to test regional Finnish populations to see how they behave in such models. Of course, Finns aren't Saamic speakers, they're Finnic speakers, and they're probably the result of a more recent Uralic expansion into Fennoscandia than the one that gave rise to the Saami.
Finnish Baltic_IA 0.671±0.076 Levanluhta_IA 0.329±0.076 chisq 14.114 tail prob 0.293508 Full outputDamgaard et al. didn't report the Y-haplogroup for Baltic_IA, but the word round the campfire is that this individual belonged to N1c, which is today the most common Y-haplogroup among Uralic speakers. Obviously, we need a lot more ancient DNA to sort all of this out, but things are already looking pretty much as expected. Stay tuned for new posts in this series following the publication of more ancient DNA relevant to this fascinating topic. See also... How did Y-haplogroup N1c get to Bolshoy Oleni Ostrov? The Uralic cline in the Global25 Late PIE ground zero now obvious; location of PIE homeland still uncertain, but...
Labels:
ancient DNA,
Bolshoy Oleni Ostrov,
Corded Ware Culture,
CWC,
Fennoscandia,
Finnic,
Finno-Ugric,
Indo-European,
Levanluhta,
N-L1026,
N1c,
Northern Europe,
Proto-Uralic,
R1a-Z645,
Saami,
Siberia,
Uralic,
Urals
Saturday, September 22, 2018
Corded Ware people =/= Proto-Uralics (Tambets et al. 2018)
A new paper on the genetic structure of Uralic-speaking populations has appeared at Genome Biology (see here). It looks to me like the prelude to a forthcoming paleogenetics paper on the same topic that was discussed in the Estonian media recently (see here). Although not exactly ground breaking (because it basically argues what I've been saying at this blog for years, like here), it's a very nice effort all round and must be read by anyone with an interest in this topic. From the paper, emphasis is mine:
Background The genetic origins of Uralic speakers from across a vast territory in the temperate zone of North Eurasia have remained elusive. Previous studies have shown contrasting proportions of Eastern and Western Eurasian ancestry in their mitochondrial and Y chromosomal gene pools. While the maternal lineages reflect by and large the geographic background of a given Uralic-speaking population, the frequency of Y chromosomes of Eastern Eurasian origin is distinctively high among European Uralic speakers. The autosomal variation of Uralic speakers, however, has not yet been studied comprehensively. Results: Here, we present a genome-wide analysis of 15 Uralic-speaking populations which cover all main groups of the linguistic family. We show that contemporary Uralic speakers are genetically very similar to their local geographical neighbours. However, when studying relationships among geographically distant populations, we find that most of the Uralic speakers and some of their neighbours share a genetic component of possibly Siberian origin. Additionally, we show that most Uralic speakers share significantly more genomic segments identity-by-descent with each other than with geographically equidistant speakers of other languages. We find that correlated genome-wide genetic and lexical distances among Uralic speakers suggest co-dispersion of genes and languages. Yet, we do not find long-range genetic ties between Estonians and Hungarians with their linguistic sisters that would distinguish them from their non-Uralic-speaking neighbours. Conclusions: We show that most Uralic speakers share a distinct ancestry component of likely Siberian origin, which suggests that the spread of Uralic languages involved at least some demic component. ... Recent aDNA studies have shown that extant European populations draw ancestry form three main migration waves during the Upper Palaeolithic, the Neolithic and Early Bronze Age [2, 3, 45]. The more detailed reconstructions concerning NE Europe up to the Corded Ware culture agree broadly with this scenario and reveal regional differences [65–67]. However, to explain the demographic history of extant NE European populations, we need to invoke a novel genetic component in Europe—the Siberian. The geographic distribution of the main part of this component is likely associated with the spread of Uralic speakers but gene flow from Siberian sources in historic and modern Uralic speakers has been more complex, as revealed also by a recent study of ancient DNA from Fennoscandia and Northwest Russia [68]. Thus, the Siberian component we introduce here is not the perfect but still the current best candidate for the genetic counterpart in the spread of Uralic languages.Citation... Tambets et al., Genes reveal traces of common recent demographic history for most of the Uralic-speaking populations, Genome Biology, (2018) 19:139 https://doi.org/10.1186/s13059-018-1522-1 See also... Big deal of 2019: ancient DNA confirms the link between Y-haplogroup N and Uralic expansions
Subscribe to:
Posts (Atom)