search this blog
Showing posts with label R1a-Z645. Show all posts
Showing posts with label R1a-Z645. Show all posts
Tuesday, July 21, 2020
The oldest R1a to date
My popular map of the oldest instances of Y-haplogroup R1a in the ancient DNA record has a new entry: PES001 from the recent Saag et al. preprint. PES001 comes from a burial site in what is now northwestern Russia and is dated to a whopping 10785–10626 calBCE.
Indeed, I'm not aware of any R1a samples older than PES001 among the treasure trove of thousands of ancient samples waiting to be published. So it's likely that this individual will remain the oldest member of our R1a clan for some years to come.
See also...
Y-haplogroup R1a and mental health
Like three peas in a pod
The mystery of the Sintashta people
Labels:
ancient DNA,
Central Asia,
Corded Ware Culture,
Eastern Europe,
Eurasia,
India,
Indo-Aryan,
Indo-Iranian,
oldest R1a,
paternal ancestry,
Pontic-Caspian steppe,
R1a,
R1a origin,
R1a-M417,
R1a-Z645,
R1a-Z93,
South Asia
Wednesday, October 16, 2019
The Battle Axe people came from the steppe (Malmstrom et al. 2019)
It's been obvious for a while now that the Corded Ware culture (CWC) and its Scandinavian variant, the Battle Axe culture (BAC), originated on the Pontic-Caspian steppe. However, Malmstrom et al. drive the point home in a new open access paper at Proceedings B [LINK]. From the paper, emphasis is mine:
The Neolithic period is characterized by major cultural transformations and human migrations, with lasting effects across Europe. To understand the population dynamics in Neolithic Scandinavia and the Baltic Sea area, we investigate the genomes of individuals associated with the Battle Axe Culture (BAC), a Middle Neolithic complex in Scandinavia resembling the continental Corded Ware Culture (CWC). We sequenced 11 individuals (dated to 3330–1665 calibrated before common era (cal BCE)) from modern-day Sweden, Estonia, and Poland to 0.26–3.24× coverage. Three of the individuals were from CWC contexts and two from the central-Swedish BAC burial ‘Bergsgraven’. By analysing these genomes together with the previously published data, we show that the BAC represents a group different from other Neolithic populations in Scandinavia, revealing stratification among cultural groups. Similar to continental CWC, the BAC-associated individuals display ancestry from the Pontic–Caspian steppe herders, as well as smaller components originating from hunter–gatherers and Early Neolithic farmers. Thus, the steppe ancestry seen in these Scandinavian BAC individuals can be explained only by migration into Scandinavia. Furthermore, we highlight the reuse of megalithic tombs of the earlier Funnel Beaker Culture (FBC) by people related to BAC. The BAC groups likely mixed with resident middle Neolithic farmers (e.g. FBC) without substantial contributions from Neolithic foragers. ... By contrast, the CWC individuals from Obłaczkowo in Poland (poz44 and poz81) show an extremely high proportion of steppe ancestry (greater than 90%), which is different from the later CWC-associated individuals excavated in Pikutkowo (Poland) [23], but similar to some other CWC-associated individuals from Germany, Lithuania, and Latvia [2,8,31]. Interestingly, these individuals with a large fraction of steppe ancestry have typically been dated to more than 2600 BCE, making them among the earliest CWC individuals genetically investigated. This observation, i.e. early CWC individuals resembled (genetically) Yamnaya-associated individuals, while later CWC groups show higher levels of European Neolithic farmer ancestry (Pearson's correlation coefficient: −0.51, p = 0.006) (figure 2), suggests an initial dispersal that occurred rapidly.See also...
Saturday, October 12, 2019
The Balkan connection
The hot topic at the moment is social inequality in Bronze Age Europe, thanks to a new paper by Mittnik et al. at Science. The full article is sitting behind an exceedingly robust paywall here.
However, the genotype dataset from the paper is freely available at the Max Planck Society's Edmond data repository here. Below is my Principal Component Analysis (PCA) of ancient West Eurasian genetic variation featuring 41 of the highest quality ancients from the new dataset. Almost all of them are from the Lech Valley in the Bavarian Alps, covering the period from the Bell Beaker culture (BBC) to the Middle Bronze Age (MBA). Two of the samples are from a mass Corded Ware culture (CWC) burial in the more northerly Tauber Valley.
I've also highlighted other ancients on the plot associated with the BBC and CWC from present-day Netherlands and Germany, respectively. The relevant PCA datasheet can be downloaded here.
Social stratification in ancient Europe is a fascinating topic, and it's an issue that I've started looking at myself (see here). However, I can't see any correlation between the inferred social standing of the individuals from the Lech and Tauber valleys and their positions in my PCA.
Nevertheless, the PCA is interesting in that it highlights considerable genetic heterogeneity within the Lech Valley BBC population. Indeed, how is this heterogeneity even possible, if, as per Mittnik et al., ancient DNA "has shown that the spread of the BBC throughout continental Europe did not involve large-scale migrations"?
Below is another version of my PCA, but this time focusing on three males: Lech Valley Beakers UNTA58_68Sk1 and WEHR_1192SkA, as well as ALT_4 from the aforementioned mass CWC grave in the Tauber Valley. Note that UNTA58_68Sk1 and WEHR_1192SkA represent genetically the most southern and northern, respectively, Lech Valley BBC samples that had enough data to be run in my analysis. I chose to focus on males because they carry the Y-chromosome, which can be informative about male-mediated ancient population expansions.
The PCA outcomes for these individuals are generally in line with their results in other types of genetic analyses, including those based on formal statistics. For instance, compared to the other two, ALT_4 harbors excess early steppe herder ancestry, UNTA58_68Sk1 excess early European farmer ancestry, and WEHR_1192SkA excess European hunter-gatherer ancestry. Moreover...
- UNTA58_68Sk1 shows a non-local isotopic signature and belongs to Y-haplogroup G2a, a marker essentially missing from BBC populations north of the Alps, and is best modeled as a two-way mixture between Bronze Age populations from the Balkans and the Pontic-Caspian steppe (see here), which probably means that he was a migrant to the Lech Valley from south of the Alps - importantly, UNTA58_68Sk1 is not an isolated case, at least in the sense that several other BBC individuals from Bavaria, Bohemia, Hungary and Poland show varying ratios of Balkan-related ancestry, although almost all of these people are women - WEHR_1192SkA is very similar to Bell Beakers from the northern Netherlands with whom he shares the R1b-P312 Y-haplogroup, suggesting that he was part of a population that moved into the Lech Valley from potentially as far away as the North Sea coast - although ALT_4 probably shares the R1b-L51 Y-haplogroup with WEHR_1192SkA and many other BBC and Bronze Age individuals from the Bavarian Alps and surrounds, this can't be used as evidence of significant local genetic continuity after the CWC period, especially considering the comparatively eastern genome-wide structure of ALT_4.Of course, archeological data suggest that the BBC was influenced in some important ways by the Copper and Bronze Age cultures of the Balkans and Carpathian Basin. So much so, in fact, that Marija Gimbutas, author of The Civilization of the Goddess, believed that the BBC originated in the Balkans from a synthesis of the local Vucedol culture and the intrusive Yamnaya culture from the Pontic-Caspian steppe. Considering the ancient DNA evidence, however, the main demographic center of the early BBC could not have been south of the Alps. Rather, it appears that early BBC and even CWC groups from north of the Alps moved into the Balkans and Carpathian Basin, where they may have established contacts with the local elites. If so, this might explain the significant southern cultural influences on the BBC, but limited accompanying genetic impact. This scenario also has support from archeological data (for instance, see here). See also... Is Yamnaya overrated? The Boscombe Bowmen Single Grave > Bell Beakers
Wednesday, September 11, 2019
Y-haplogroup R1a and mental health
I've updated my map of pre-Corded Ware culture R1a samples with a couple of new entries from Central and South Asia (the original is still here). However, before any of you get overly excited, please note that these samples aren't older than the Corded Ware culture. The reason I added them to my map is to counter the ongoing absurd claims online that South Asian R1a isn't derived from European R1a.
Just in case the map can't be viewed in all of its glory in some devices, here's what the fine print says:
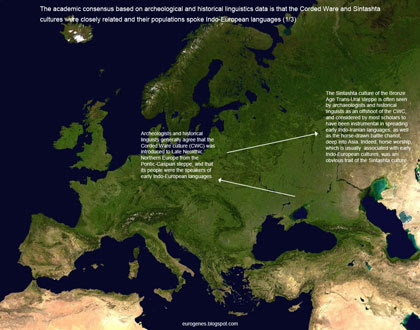
The oldest example of R1a in ancient DNA from Central Asia is dated to 2132-1940 calBCE (ID I3770, Narasimhan 2019). Moreover, this sequence is closely related to much older R1a samples from Central, Eastern and Northern Europe, and phylogenetically nested within their diversity. Thus, it must surely represent a population expansion from Europe to Central Asia. Indeed, it's also associated with the Bronze Age Andronovo archeological culture, which is usually seen as an offshoot of the Corded Ware culture (CWC) of Late Neolithic Europe. The vast majority of present-day R1a lineages in Central Asia are closely related to that of I3770, and so must also ultimately derive from Europe. The oldest instance of R1a in ancient DNA from South Asia is dated to just 1044-922 calBCE (ID I12457, Narasimhan 2019). This sequence, as well as the vast majority of present-day South Asian R1a lineages, are closely related to much older R1a samples from Central, Eastern and Northern Europe, and phylogenetically nested within their diversity. Thus, they must surely represent a population expansion from Europe to South Asia via Central Asia, in all likelihood during the Bronze Age. Even if R1a existed in South Asia before the Bronze Age, which is extremely unlikely, because it's found in samples from indigenous European hunter-gatherers, the vast majority of present-day R1a lineages in South Asia must be ultimately from Europe.The idea that most, if not all, South Asian R1a is derived from European R1a seriously scares a lot of people. This is obvious in many online discussions on the topic. I suspect they're so frightened by it because, in their minds, it has the potential to encourage discrimination and even racism, perhaps by re-defining the colonization of much of the world by European nations in the recent past as the natural order of things? In any case, clearly we're dealing with some sort of mass phobia here. I've got advice for those of you suffering from this problem: if you're honestly worried that the geographic provenance and expansion history of some Y-haplogroup is going to negatively impact on your life in any meaningful way, then it's time to find yourself a quality mental health professional. All the best with that. See also... The mystery of the Sintashta people The Poltavka outlier Yamnaya isn't from Iran just like R1a isn't from India
Labels:
ancient DNA,
Central Asia,
Corded Ware Culture,
Eastern Europe,
Eurasia,
India,
Indo-Aryan,
Indo-Iranian,
oldest R1a,
paternal ancestry,
Pontic-Caspian steppe,
R1a,
R1a origin,
R1a-M417,
R1a-Z645,
R1a-Z93,
South Asia
Saturday, June 15, 2019
Not Bell Beaker, not Corded Ware, but...the SGBR complex
I'd be very grateful if someone could explain to me what this new paper at the Proceedings of the Prehistoric Society journal was actually about.
Citation...
Furholt, Martin, Re-integrating Archaeology: A Contribution to aDNA Studies and the Migration Discourse on the 3rd Millennium BC in Europe, Proceedings of the Prehistoric Society, Published online: 10 June 2019, DOI: https://doi.org/10.1017/ppr.2019.4
See also...
Tuesday, May 7, 2019
The execution
Around 2,800 BCE, in what is now southern Poland, a family group of fifteen individuals associated with the Globular Amphora culture (GAC) were massacred. They were probably captured and executed, because each victim was killed with a blow to the head from the same type of weapon, possibly a stone axe, and lacked defensive wounds. The dead were mostly women and children. They were buried in a mass grave, but with great care and very likely by someone who knew them well.
This Late Neolithic mass grave is the focus of a new ancient DNA and archeological research paper at PNAS by Schroeder et al. (see here). The authors tentatively attribute the massacre to the Corded Ware culture (CWC) people, who were expanding rapidly at the time across much of Europe from their homeland on the Pontic-Caspian steppe.
The CWC people may or may not have been responsible; we'll never know for sure. The perpetrators could just as easily have been a competing GAC family group.
In any case, it's interesting to see that the GAC males belong to Y-chromosome haplogroup I2a-L801. This is today a rather uncommon subclade of I2, and almost exclusively found in Germanic-speaking populations, especially Scandinavians. To me this suggests that some Polish GAC males were incorporated into Indo-European-speaking CWC populations that ended up in Scandinavia, and their paternal lineages eventually became a part of the Proto-Germanic gene pool. Admittedly, though, that's just one of many possible scenarios.
See also...
Late PIE ground zero now obvious; location of PIE homeland still uncertain, but...
Corded Ware people =/= Proto-Uralics (Tambets et al. 2018)
Inferring the linguistic affinity of long dead and non-literate peoples: a multidisciplinary approach
Labels:
ancient DNA,
Corded Ware Culture,
execution,
GAC,
Germanic,
Globular Amphora,
I2a,
Indo-European,
Late Neolithic,
mass burial,
mass grave,
massacre,
Poland,
Pontic-Caspian steppe,
R1a-M417,
R1a-Z645,
stone axe
Sunday, May 5, 2019
Conan the Barbarian probably belonged to Y-haplogroup R1a
A fresh batch of Iron Age genomes from across the Eurasian steppes is about to be published along with a new paper at Current Biology. The manuscript, titled Shifts in the Genetic Landscape of the Western Eurasian Steppe Associated with the Beginning and End of the Scythian Dominance, is still under review but freely available here.
Most of the male ancients, including two Cimmerians from the North Pontic steppe, in what is now Ukraine, belong to Y-chromosome haplogroup R1a. Wasn't Conan the Barbarian supposed to be a Cimmerian? From the preprint, emphasis is mine:
The Early Iron Age nomadic Scythians have been described as a confederation of tribes of different origins, based on ancient DNA evidence [1-3]. It is still unclear how much of the Scythian dominance in the Eurasian Steppe was due to movements of people and how much reflected cultural diffusion and elite dominance. We present new whole-genome sequences of 31 ancient Western and Eastern Steppe individuals including Scythians as well as samples pre- and postdating them, allowing us to set the Scythians in a temporal context (in the Western/Ponto-Caspian Steppe). We detect an increase of eastern (Altaian) affinity along with a decrease in Eastern Hunter-Gatherer (EHG) ancestry in the Early Iron Age Ponto- Caspian gene pool at the start of the Scythian dominance. On the other hand, samples of the Chernyakhiv culture postdating the Scythians in Ukraine have a significantly higher proportion of Near Eastern ancestry than other samples of this study. Our results agree with the Gothic source of the Chernyakhiv culture and support the hypothesis that the Scythian dominance did involve a demic component. ... Out of the 31 samples of this study, 16 are male, and with sufficient Y-chromosome coverage for haplogroup assignment (Table S2). R1a (43%) and I (27%) are the two most frequent Y- chromosome hgs in present-day Ukrainians [142]. R1a is also the predominant lineage among Cimmerians, Scy_Ukr and ScySar_SU in our data, and present among Scy_Kaz as well. Thus, although acknowledging our small sample size, the individuals sampled from archaeological context associated with Scythian identity do not appear to stand out from the context of other groups living in the region before and after them. One notable difference from the present is the absence of hg N, nowadays widespread in the Volga-Uralic region and West Siberia as well as among Mongols and Altaians [165-167]; however, this result is consistent with the absence of hg N among Bronze Age and Eneolithic males from the Steppe [168]. In context of their claimed Altaian homeland it is interesting to note that one Scy_Ukr and the single Sar_Cau sample belong to the Q1c-L332 lineage which is a sub-clade of hg Q1c-L330 that today has peak frequency of 68% in Western Mongolians [169] and occurs at 17% in South Altaians [170] while being very rare (<1%) in East European populations and absent elsewhere (https://www.yfull.com/tree/Q-L330/).Järve et al., Shifts in the Genetic Landscape of the Western Eurasian Steppe Associated with the Beginning and End of the Scythian Dominance, Current Biology (preprint), Posted: 6 Mar 2019, http://dx.doi.org/10.2139/ssrn.3346985 Update 12/07/2019: The paper has just been published and is freely available at Current Biology [LINK]. See also... The mystery of the Sintashta people On the association between Uralic expansions and Y-haplogroup N Late PIE ground zero now obvious; location of PIE homeland still uncertain, but...
Labels:
ancient DNA,
Barbarian,
Chernyakhiv culture,
Chernyakhov culture,
Cimmerian,
Eurasian steppe,
Germanic,
Gothic,
Indo-European,
Indo-Iranian,
Iron Age,
North Pontic,
R1a-M417,
R1a-Z645,
Sarmatian,
Scythian,
Thracian
Thursday, April 25, 2019
Some myths die hard
Ancient DNA tells us that the Bronze Age wasn't kind to the indigenous populations of Central Asia. It seems to have wiped them out totally. Indeed, Central Asia might well be the only major world region in which native hunter-gatherers failed to make a perceptible impact on the genetics of any extant populations.
Before the Neolithic transition, much of Central Asia was home to hunter-gatherers closely related to those of nearby western Siberia. During the Neolithic, agriculturalists and pastoralists from the Near East gradually moved into the more arable parts of southern and eastern Central Asia, eventually giving rise to the Bactria Margiana Archaeological Complex, or BMAC, and other similar communities.
It's not clear what their relationship was like with the native hunter-gatherers in these areas. But they did mix with them in varying degrees. This is obvious because genome-wide genetic ancestry characteristic of the Botai people, who hunted and eventually domesticated horses on the Kazakh steppe during the 4th millennium BCE, and were probably the archetypal Central Asians for their time, is found at significant levels in a number of later samples from Central Asian farmer and pastoralist sites, such as Dali, Gonur Tepe and Sarazm.
Thus, even though the Neolithic transition did have a big impact on Central Asia, and clearly led to large scale population replacements in some parts of the region, this was just the beginning of these population shifts. Moreover, in some cases the expanding farmer and pastoralist populations seem to have acquired significant indigenous Central Asian ancestry and spread it with them.
The precise geographic extent of the relatively unique Botai-related ancestry in prehistoric Eurasia is still something of a mystery. But to give you a general picture of where it was found from around 6,000 BCE to 2,000 BCE, here's a map with info about samples with significant levels of this type of ancestry from a wide range of sites in space and time.
Going by this map, I'd say it's safe to infer that the Botai-related ancestry was a major feature of practically all forager populations living between the Caspian Sea and the Altai Mountains. It was also present in the Early Bronze Age (EBA) pastoralist population associated with the Steppe Maykop archeological culture of Eastern Europe, so it may have already been in Europe as early as 3,800 BCE, because that's when the Steppe Maykop culture first appeared.
It's an interesting question where the ancestors of the Steppe Maykop herders came from. I once simply assumed that they were closely related to the Maykop people who lived in the Caucasus Mountains. But it's now clear that the populations associated with these two similar cultures were starkly different, with the Maykop people being basically of Near Eastern origin and lacking any discernible Botai-like ancestry. My guess for now is that the Steppe Maykop herders were in large part the descendants of the Kelteminar culture population from just east of the Caspian Sea, but we'll see about that when more ancient DNA comes in.
The other great mystery is what eventually happened to the Steppe Maykop people. Around 3,000 BCE, their culture vanished from the archeological record and their particular genetic signature disappeared from the steppe ancient DNA record. Where did they go? Did they migrate back east?
I don't know, but at about that time other Eastern European steppe herders, those associated with the Yamnaya and Corded Ware archeological cultures, began to stir and migrate in big numbers in basically all directions, including into Steppe Maykop territory. Indeed, unlike the Steppe Maykop population, these groups weren't closely related to any contemporaneous or earlier Central Asians. But they ended up moving into Central Asia, and in a big way too.
Their impact all the way from the Ural Mountains to what are now China and India was profound. For instance, not only did they end up totally replacing the Botai people, but also their horses. For more details on this topic check out the Youtube clip here. I have a strong suspicion that the same sort of thing happened to the aforementioned Steppe Maykop people. In other words, they may have been forced out from the Eastern European steppe, and perhaps sought shelter in the Caucasus Mountains?
Admittedly, I'm not offering anything new here. I just wanted to emphasize a few key points, because I'm still seeing some confusion online about the population history of Central Asia, and especially how it relates to the population history of Europe, and also the Proto-Indo-European homeland question. Make no mistake, thanks to the ancient DNA already available from Central Asia, we can confidently infer the following:
- the chance that the ancient European populations associated with the Yamnaya, Corded Ware and other closely related archeological cultures formed as a result of migrations from Central Asia is zero - the chance that the Proto-Indo-European homeland was located in Central Asia is zero - the chance that present-day Europeans, by and large, derive from any ancient Central Asian populations is zeroSee also... Central Asia as the PIE urheimat? Forget it The Steppe Maykop enigma Late PIE ground zero now obvious; location of PIE homeland still uncertain, but...
Labels:
Afanasievo,
ancient DNA,
Andronovo,
Botai,
Central Asia,
Corded Ware Culture,
Eastern Europe,
horse,
India,
Pontic-Caspian steppe,
Proto-Indo-European,
R1a-M417,
R1a-Z645,
R1a-Z93,
R1b-M269,
Sintashta,
wheel,
Yamnaya
Sunday, March 31, 2019
Map of pre-Corded Ware culture (>2900 BCE) instances of Y-haplogroup R1a (updated)
Below is a map showing the global distribution of Y-chromosome haplogroup R1a prior to the expansions of the R1a-rich Corded Ware culture (CWC) people and their descendants across Europe and Asia from around 2900 BCE. I'll be updating this map regularly and using it to help me narrow down the options for the place of origin of R1a, and also to counter the misinformation about this topic that has appeared in print and online over the years, including in many scientific publications and popular websites such as Wikipedia.
Incredibly, as far as I know, there are just six reliably called instances of R1a in the now ample Eurasian ancient DNA record dating to the pre-CWC period. To put this into perspective, consider that R1a is today the most common Y-haplogroup in much of Europe and Asia. How did that happen I wonder? However, please note that I chose to base the map only on samples sequenced with the capture and shotgun methods, rather than the PCR method, which is susceptible to producing contaminated results and no longer used in major ancient DNA studies.
See also...
Y-haplogroup R1a and mental health
The Poltavka outlier
Late PIE ground zero now obvious; location of PIE homeland still uncertain, but...
Labels:
ancient DNA,
Bronze Age,
Copper Age,
Corded Ware Culture,
CWC,
Eastern Europe,
Eurasia,
PIE,
Proto-Indo-European,
R1a,
R1a origin,
R1a-M17,
R1a-M198,
R1a-M417,
R1a-M420,
R1a-Z645,
R1a-Z93,
R1a1a1,
Y-haplogroup
Monday, December 3, 2018
On the trail of the Proto-Uralic speakers (work in progress)
Historical linguists have long posited that Fennoscandia was a busy contact zone between early Germanic and Uralic languages. The first ancient DNA samples from what is now Finland have corroborated their inferences, by showing that during the Iron Age the western part of the country was inhabited by a genetically heterogeneous population closely related to both the Uralic-speaking Saami and Germanic-speaking southern Scandinavians.
The samples were sequenced and analyzed by two different teams of researches, and their findings published recently in Lamnidis et al. and Sikora et al. (see here and here, respectively).
This is how most of these ancients, whose remains were excavated from the Levanluhta burial site dated to 300–800 CE, behave in a Principal Component Analysis (PCA) based on my Global25 data. Levanluhta_IA are the Saami-related samples, while Levanluhta_IA_o is an Scandinavian-like outlier. Baltic_IA is an Iron Age individual from what is now Lithuania from the recent Damgaard et al. paper (see here). Note the accuracy of the Global25 data in pinpointing their genetic affinities and also the trajectory of the Levanluhta_IA cluster, which seems to be "pulling" towards Levanluhta_IA_o.
The Saami and Levanluhta_IA are clear outliers from the main Northern European cluster. There are two reasons for this: excess East Asian/Siberian-related ancestry and Saami-specific genetic drift. However, this eastern admixture and genetic drift are shared in varying degrees by other North European populations, especially those that also speak Uralic languages, and this is why they appear to be "pulling" towards the Saami/Levanluhta_IA clusters in my PCA. Thus, what this suggests is that the expansion of Uralic languages across Northeastern Europe was intimately linked with the spread of Siberian-related ancestry into the region.
This idea has been around for a long time and is now becoming even more widely accepted (see here). However, Lamnidis et al. also featured samples from a likely pre-Uralic (1523±87 calBCE) burial site at Bolshoy Oleni Ostrov in the Kola Peninsula, present-day northern Russia, and, perhaps surprisingly, found that they showed even more Siberian-related ancestry than Levanluhta_IA. So what's going on?
I'm confident that this discrepancy can be explained by multiple waves of migrations from the east into Northeastern Europe, possibly before, during and after the time of the people buried at Bolshoy Oleni Ostrov, by pre-Uralic, para-Uralic and/or Proto-Uralic-speaking populations.
Consider the following qpAdm output, in which Levanluhta_IA is just barely modeled successfully as a two-way mixture between Levanluhta_IA_o and Bolshoy_Oleni_Ostrov. The statistical fit improves significantly with the addition of Glazkovo_EBA as a third mixture source. This is an ancient population from near Lake Baikal dated to 4597-3726 BC from the aforementioned Damgaard et al. paper.
Levanluhta_IA Bolshoy_Oleni_Ostrov 0.468±0.036 Levanluhta_IA_o 0.532±0.036 chisq 19.129 tail prob 0.0854706 Full output Levanluhta_IA Bolshoy_Oleni_Ostrov 0.241±0.092 Glazkovo_EBA 0.162±0.059 Levanluhta_IA_o 0.597±0.046 chisq 7.756 tail prob 0.734966 Full outputFor the sake of being complete, I also tested whether Levanluhta_IA_o could be substituted by other similar ancient samples from the neighborhood, including those associated with the Battle-Axe and Corded Ware cultures. There's not much to report; qpAdm returned poor statistical fits and/or implausible ancestry proportions (for the full output from my runs, see here). Baltic_IA did produce a statistically sound model, but with excess Glazkovo_EBA-related ancestry. I also had to drop Bolshoy_Oleni_Ostrov from the analysis to make things work, which suggests to me that the result shouldn't be taken too literally.
Levanluhta_IA Baltic_IA 0.677±0.034 Glazkovo_EBA 0.323±0.034 chisq 8.547 tail prob 0.741095 Full outputSo as far as I can see, the western ancestry in Levanluhta_IA is likely to be mostly of Germanic origin, and thus Indo-European, meaning that it's logical to look east, perhaps far to the east, for the source of its Uralic ancestry. This might seem like a complicated and uncertain task, considering that Levanluhta_IA could well be at least a thousand years younger than the first entry of Uralic speakers into Fennoscandia. However, take a look what happens when I substitute Glazkovo_EBA with a variety of Uralic-speaking populations from around the Ural Mountains, which is where the Proto-Uralic homeland is generally considered to have been located.
Levanluhta_IA Bolshoy_Oleni_Ostrov 0.210±0.091 Khanty 0.283±0.090 Levanluhta_IA_o 0.507±0.035 chisq 7.007 tail prob 0.798532 Full output Levanluhta_IA Bolshoy_Oleni_Ostrov 0.193±0.098 Levanluhta_IA_o 0.495±0.035 Mansi 0.312±0.100 chisq 7.884 tail prob 0.7237 Full output Levanluhta_IA Bolshoy_Oleni_Ostrov 0.300±0.065 Levanluhta_IA_o 0.337±0.072 Mari 0.363±0.121 chisq 8.393 tail prob 0.677705 Full output Levanluhta_IA Bolshoy_Oleni_Ostrov 0.238±0.084 Levanluhta_IA_o 0.553±0.036 Nenets 0.209±0.067 chisq 7.210 tail prob 0.78181 Full output Levanluhta_IA Bolshoy_Oleni_Ostrov 0.302±0.069 Levanluhta_IA_o 0.324±0.081 Udmurt 0.373±0.135 chisq 9.195 tail prob 0.60393 Full outputAll of these models look great, and easily rival the best model with Glazkovo_EBA. Moreover, they make good sense in terms of linguistics. The only problem is that they're anachronistic, because the Uralic-speaking reference populations are younger than Levanluhta_IA. So I can't be certain that they reflect reality without corroboration from ancient DNA. It might turn out, for instance, that a Glazkovo_EBA-like population was already present somewhere deep in Europe before or during the time of Bolshoy_Oleni_Ostrov, while no such population existed around the Ural Mountains until the time of Levanluhta_IA. By the way, it might be important to note that the present-day Finnish samples in my dataset can't be modeled as a mixture between Levanluhta_IA and Levanluhta_IA_o. But they can be modeled as a mixture between Baltic_IA and Levanluhta_IA. I don't know which part of Finland they're from exactly; probably all over the place, so it'd be useful to test regional Finnish populations to see how they behave in such models. Of course, Finns aren't Saamic speakers, they're Finnic speakers, and they're probably the result of a more recent Uralic expansion into Fennoscandia than the one that gave rise to the Saami.
Finnish Baltic_IA 0.671±0.076 Levanluhta_IA 0.329±0.076 chisq 14.114 tail prob 0.293508 Full outputDamgaard et al. didn't report the Y-haplogroup for Baltic_IA, but the word round the campfire is that this individual belonged to N1c, which is today the most common Y-haplogroup among Uralic speakers. Obviously, we need a lot more ancient DNA to sort all of this out, but things are already looking pretty much as expected. Stay tuned for new posts in this series following the publication of more ancient DNA relevant to this fascinating topic. See also... How did Y-haplogroup N1c get to Bolshoy Oleni Ostrov? The Uralic cline in the Global25 Late PIE ground zero now obvious; location of PIE homeland still uncertain, but...
Labels:
ancient DNA,
Bolshoy Oleni Ostrov,
Corded Ware Culture,
CWC,
Fennoscandia,
Finnic,
Finno-Ugric,
Indo-European,
Levanluhta,
N-L1026,
N1c,
Northern Europe,
Proto-Uralic,
R1a-Z645,
Saami,
Siberia,
Uralic,
Urals
Saturday, September 22, 2018
Corded Ware people =/= Proto-Uralics (Tambets et al. 2018)
A new paper on the genetic structure of Uralic-speaking populations has appeared at Genome Biology (see here). It looks to me like the prelude to a forthcoming paleogenetics paper on the same topic that was discussed in the Estonian media recently (see here). Although not exactly ground breaking (because it basically argues what I've been saying at this blog for years, like here), it's a very nice effort all round and must be read by anyone with an interest in this topic. From the paper, emphasis is mine:
Background The genetic origins of Uralic speakers from across a vast territory in the temperate zone of North Eurasia have remained elusive. Previous studies have shown contrasting proportions of Eastern and Western Eurasian ancestry in their mitochondrial and Y chromosomal gene pools. While the maternal lineages reflect by and large the geographic background of a given Uralic-speaking population, the frequency of Y chromosomes of Eastern Eurasian origin is distinctively high among European Uralic speakers. The autosomal variation of Uralic speakers, however, has not yet been studied comprehensively. Results: Here, we present a genome-wide analysis of 15 Uralic-speaking populations which cover all main groups of the linguistic family. We show that contemporary Uralic speakers are genetically very similar to their local geographical neighbours. However, when studying relationships among geographically distant populations, we find that most of the Uralic speakers and some of their neighbours share a genetic component of possibly Siberian origin. Additionally, we show that most Uralic speakers share significantly more genomic segments identity-by-descent with each other than with geographically equidistant speakers of other languages. We find that correlated genome-wide genetic and lexical distances among Uralic speakers suggest co-dispersion of genes and languages. Yet, we do not find long-range genetic ties between Estonians and Hungarians with their linguistic sisters that would distinguish them from their non-Uralic-speaking neighbours. Conclusions: We show that most Uralic speakers share a distinct ancestry component of likely Siberian origin, which suggests that the spread of Uralic languages involved at least some demic component. ... Recent aDNA studies have shown that extant European populations draw ancestry form three main migration waves during the Upper Palaeolithic, the Neolithic and Early Bronze Age [2, 3, 45]. The more detailed reconstructions concerning NE Europe up to the Corded Ware culture agree broadly with this scenario and reveal regional differences [65–67]. However, to explain the demographic history of extant NE European populations, we need to invoke a novel genetic component in Europe—the Siberian. The geographic distribution of the main part of this component is likely associated with the spread of Uralic speakers but gene flow from Siberian sources in historic and modern Uralic speakers has been more complex, as revealed also by a recent study of ancient DNA from Fennoscandia and Northwest Russia [68]. Thus, the Siberian component we introduce here is not the perfect but still the current best candidate for the genetic counterpart in the spread of Uralic languages.Citation... Tambets et al., Genes reveal traces of common recent demographic history for most of the Uralic-speaking populations, Genome Biology, (2018) 19:139 https://doi.org/10.1186/s13059-018-1522-1 See also... Big deal of 2019: ancient DNA confirms the link between Y-haplogroup N and Uralic expansions
Subscribe to:
Posts (Atom)