The definition of Sredni Stog phenomenon (SSP) varies and is often a loosely applied term to refer to pretty much any individual in the Dnieper and Don regions between the ‘Neolithic’ and Yamnaya periods. In order to elucidate the SSP, some brief remarks on the preceding Mariupol phenomenon are warranted. Understanding the Mariupol horizon is fairly straightforward – its development was catalysed by an intrusion of groups from somewhere west of the Dnieper, ~ 5500BC. The ‘proto-Mariupol’ group were genomic and economic ‘hunter-gatherers’, lacking any discernible EEF admixture, and with Y-hg I-L702 uniparental ‘trace-dye’. The Mariupol phenomenon predominantly impacted the lower reaches of the Dnieper and Azov steppe (Lower Dnieper- Azov group, ‘LDA’), but extended toward the Don, Volga and even the Kuban steppe in an attenuated form. The elevated levels of “Ukr N’ in Golubaya Krinitsa and the Y-hg I2a-L702 individual at Berezhnovka attest to this movement. The Mariupol phenomenon is associated with the development of formal cemeteries, linking them with Late Neolithic mixed farmer/HG groups in the northeast Balkans. Individuals were buried in a ‘supine straight-legged’ inhumation, with grave goods such as boar-tusk pendants for select males and adorning shell beads for females. This might signal the emergence of gender-differentiation in burials and the rise of local leaders or ‘chiefs’. Data sets treat the ‘Neolithic’ in Ukraine as a monolithic phenomenon, however it is important to note that Dereivka stands apart – it is 200km north of other “Ukraine_N” sites such as Volnienski, Igren and Vovnigi, beyond the ‘Dnipro bend’. Moreover, male individuals from Dereivka are overwhelmingly assigned to Y-hg R1b-V88 and the burial pose (N-S) at Dereivka deviates from the more common E-W orientation seen elsewhere. Quite a few of the published ‘Sredni Stog’ individuals are from Dereivka, and often earlier than 4500BC, and N. Kotova assigns them to the Dnieper-Donets culture. Moreover, the recently published middle Don individuals, such as those from Golubaya Krinitsa and Vasilyevsky Cordon -17, are also not Sredni Stog, but can be thought of as ‘partially Mariupolised hunter-gatherers’. In another example, the (undated) ‘Sredni Stog’ individual I27930 from Igren was assigned to Y-hg Q and he can be modelled as a 2-way mix of EHG & WHG. This individual is actually from the Mesolithic. So what occurred during the Sredni Stog period? In contrast to the Mariupol phase, the population dynamics associated with SSP are complex: at least three external flows can be highlighted (i) the advance of Tripolje communities from the Carpathians to the Dnieper (ii) arrival of South-Caucasian/CHG agro-pastoralists in the north Caucasus, and (iii) arrival of ‘central Asian’ populations in Volga-Caspian region (represented by “TTK individual’); in addition to intra-steppe shifts and flows. Notwithstanding, the ‘ideological background’ of SSP is rooted in the Mariupol horizon. The stereotypical SSP burials features individuals buried on their back, but increasingly with legs up-flexed. And we see the beginnings of kurgan constructions, which vary from stone cairns to soil-thrown barrows. Most are buried in simple pits, however some have more complex ‘catacomb’ pits. What happened in the Dniester-Dnieper-Don region during the SSP? We can begin by orientating ourselves with a PCA to observe two main clines developing. One cline develops between ‘Ukr_N and EEF and a second cline pulls toward Lower-Volga Caucasus groups. The first cline mostly comprises of ‘Farmers’ from Tipolje and ‘hunter-gatherers’ from Dereivka. The second cline consists of individuals from Dereivka and the lower Dnieper-Azov group pulling toward Lower Volga-Caucasus groups. Admixture analysis with qpAdm reveals 3 groups within the 2 broad clines. The first group can be thought of as ‘core Sredni Stog’. These individuals are 2-way mixes of ‘Ukr_N’ and ‘Steppe Eneolithic’ (sometimes Progress works, sometimes Remontoye or Berezhnovka). They are both males and females. In our examples, the females are from Kopachiv Yar (4000 BC) and Dereivka (3500BC). The males come from Dereivka (4300BC), Moluykhiv Bugor (4000BC), Vynohrado (4000BC); they are all derived for Y-hg I2a-L703+. These results represent a blending of social networks between the LDA and various lower Volga-Caucasus groups, and the subsequent expansion by LDA further West. The terminus ante quem of 4300 BC matches the corrected dating of the Kuban steppe sites such as Progress & Vojnuchka. Another subset comprises of individuals from Dereivka and Verteba cave who situated on an ‘Ukr-N’ < - - > EEF cline. Many of the earlier Dereivka individuals are almost 100% Ukr_N. Verteba cave Tripolje can be modelled as 80% EEF + 20% Ukr_N. One individual from Dereikva (I3719) falls outside the Dereivka <-> Tripolje clin, as he plots further ‘south’ with Balkan-LBK farmers. Consistently, he comes out as ~100% EEF with qpAdm. Dating to ~4700BC, he precedes the arrival of Tipolje groups to the region by hundreds of years. FtDNA have assigned him to I2-Z161- FTH81, which is distinctive to the LDA haplotypes and is phylogenetically linked to a Czech LBK individual. A third group consists of individuals with more complex 3-way ancestries, consisting of EEF, Ukr-N and Steppe_En and/or Maikop. These come from late Dereivka and late Tripolje groups, in archaeological literature often termed as ‘Soldanesti’, ‘Zhivotilovksa-Volchansk’, Cernavoda (Kartal). Once again, the males from Soldanesti and Cernavoda derive from LDA-related Y-hg I2a-L703 in some shape or form. Conclusions: 1) Firstly, we note that the Dereivka group was subject to early EEF influence, as soon as eastern LBK groups reached Ukraine after 5000BC. However, their main interaction occurred with the younger, Tripolje group, which expanded toward the Dnieper after ~ 4300 BC. 2) In the LDA group we observe patrilineal continuity. These clans created expansive social-networks. They initially mixed with groups from the lower Volga-Caucasus area. Some then moved west, and ‘took over’ the Tripolje region and acquired high levels of EEF. 3) As a third conclusion, we can reject the commonly held notion that Tripolje was ‘conquered by Yamnaya pastoralists’. Our analysis instead highlights that their core structure fragmented as they became intertwined with powerful networks to their west (Trpolje) and east (Sredni Stog). The ‘take-over’ was due to the expansion of LDA/ SS groups. Mixed groups emerged such as Cernavoda and Soldanesti, which retained Tripolje ancestry and some cultural traditions. By the time Yamnaya groups reached the Dniester forest steppe, Tripolje had been long gone.See also... ‘Proto-Yamnaya’ Eneolithic individuals from Kuban steppe c. 3700 BC ? (guest post)
search this blog
Showing posts with label Eastern Europe. Show all posts
Showing posts with label Eastern Europe. Show all posts
Tuesday, August 12, 2025
Tripolje, Dereivka, and the Sredni Stog phenomenon (guest post)
This is another guest post by an anonymous contributor. Again, I don't necessarily agree with the author, but is he wrong? Feel free to let me know in the comments below.
Wednesday, December 4, 2024
The PIE homeland controversy: December 2024 open thread
It seems like we're getting close to the moment when Iosif Lazaridis has to finally admit that the Proto-Indo-European (PIE) homeland was located in Eastern Europe, and also that the ancestors of the Hittites and other Anatolian speakers entered Anatolia via the Balkans.
Let's discuss.
However, please note that comments from total morons, trolls and/or mentally unstable people will not be approved.
See also...
Indo-European crackpottery
Labels:
Anatolia,
ancient DNA,
Balkans,
Dear Iosif,
Eastern Europe,
I-L699,
I2a,
I2a-L699,
Indo-Anatolian,
Iosif Lazaridis,
North Pontic,
North Pontic steppe,
Pontic-Caspian steppe,
Proto-Indo-European
Monday, March 25, 2024
High-resolution stuff
I just emailed this to the authors of High-resolution genomic ancestry reveals mobility in early medieval Europe, a new preprint at bioRxiv [LINK].
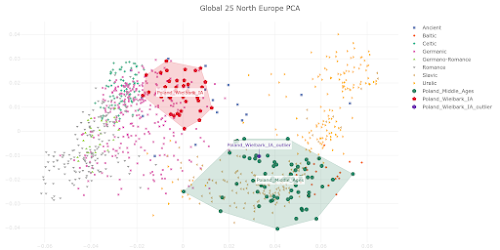
I appreciate that Polish population history is not the main focus of your preprint, and also that you're constrained by the lack of relevant and suitably high quality ancient genomes from East-Central and Eastern Europe. However, I must say that your analysis of the Medieval Polish population and resulting conclusions about Polish population history don't reflect reality. Your Poland_Middle_Ages genomic cluster is made up of just six samples that don't fully represent the genetic complexity of the core population of Medieval Poland. As a result, you classified PCA0148 as one of the Poland_Middle_Ages outliers, even though this sample isn't an outlier when analyzed within the context of the full set of published Polish Medieval genomes. Moreover, PCA0148 is very similar to several Polish Viking Age samples that show Scandinavian-specific genome-wide and Y-chromosome haplotypes, and probably likewise shows some Scandinavian-related ancestry. This is important to note when attempting to recapitulate Polish population history, because it suggests that Scandinavian-related ancestry played a formative role in the shaping of the core Polish Medieval genetic cluster. Thus, you might be correct when you claim that the six samples in your Poland_Middle_Ages cluster don't show any "detectable" Scandinavian-related ancestry, but this doesn't necessarily mean that this type of ancestry isn't a key part of the post-Iron Age Polish population history. Below is a self-explanatory Principal Component Analysis (PCA) plot that illustrates my points. Interestingly, Figure 3c in your preprint shows very similar outcomes in regards to the post-Iron Age Polish population history. But the style and scale of your figure makes it difficult to spot the subtle but likely genuine Northwest European-related genetic shifts shown by PCA0148, the Viking context samples and present-day Poles relative to the Poland_Middle_Ages cluster. However, I'm also skeptical that your Poland_Middle_Ages cluster doesn't carry any detectable or even significant Scandinavian-related ancestry. That's because I suspect that there might be some technical issues with your analysis that are masking this type of ancestry in the Polish samples. Your top mixture model for the Poland_Middle_Ages cluster is, in all likelihood, an extreme statistical abstraction of reality, rather than a close reflection of it. That's because, due to a combination of historical, geographical and genetic factors, neither Italy.Imperial(I).SG nor Lithuania.IronRoman.SG are realistic formative source populations for the Medieval Polish gene pool. One of the reasons why you ended up with such a surprising result is probably the lack of suitable samples from East-Central and Eastern Europe, especially those associated with plausibly the earliest Slavic-speaking populations. It's also possible that basing your mixture model on formal statistics played a key part. Formal statistics-based mixture models are known to be biased towards outcomes involving mixture sources from the extremes of mixture clines. If your analysis is affected by this problem, then this would help to explain why you characterized the Poland_Middle_Ages cluster as simply a two-way mixture between a Middle Eastern-related group from Imperial Rome and a Baltic population with a very high cut of European hunter-gatherer ancestry. I do note that on page 6 of your manuscript you consider the possibility that the Southern European-related signal in the Poland_Middle_Ages cluster might only be very distantly related to Italy.Imperial(I).SG, and that it may even have spread across Poland with early Slavic speakers. This is a great point, and I think it should be emphasized and expanded upon, because I suspect that the problem runs deeper than this. For instance, if the early Slavic ancestors of Poles carried substantially more Southern European-related ancestry than Lithuania.IronRoman.SG, and this ancestry was, say, more Balkan-related than Italian-related, then this might radically change your modeling of the Poland_Middle_Ages cluster. That's because these early Slavs would be positioned in a very different genetic space than Lithuania.IronRoman.SG, which could potentially require a significant signal of Scandinavian-related ancestry to get a robust mixture model. Finally, it might be useful to consider Isolation-by-Distance as a partial vector for the Italy.Imperial(I).SG-related signal in Medieval Poland. The full set of published Polish Medieval genomes includes a number of outliers with obvious ancestry from Western Europe and the Balkans. These people probably don't represent any large-scale migrations into Poland, but rather the movements of individuals and small groups. Over time, such small-scale mobility may have had a fairly significant impact on the genetic character of the Polish population.Update 26/03/2024: I sent another email to Speidel et al., this time in regards to their analysis of present-day Hungarians.
Your preprint also claims that present-day Hungarians are genetically similar to Scythians, and that this is consistent with the arrival of Magyars, Avars and other eastern groups in this part of Europe. However, present-day Hungarians are overwhelmingly derived from Slavic and German peasants from near Hungary. This is not a controversial claim on my part; it's backed up by historical sources and a wide range of genetic analyses. Hungarians still show some minor ancestry from Hungarian Conquerors (early Magyars), but this signal only reliably shows up in large surveys of Y-chromosome samples. The Scythians that you used to model the ancestry of present-day Hungarians are of local, Pannonian origin, and they don't show any eastern nomad ancestry. So they're either acculturated Scythians, or, more likely, wrongly classified as Scythians by archeologists. And since these so-called Scythians lack eastern nomad ancestry, the similarity between them and present-day Hungarians is not a sign of the impact from Avars, Hungarian Conquerors and the like, but rather a lack of significant input from such groups in present-day Hungarians.Citation... Speidel et al., High-resolution genomic ancestry reveals mobility in early medieval Europe, bioRxiv, Posted March 19, 2024, doi: https://doi.org/10.1101/2024.03.15.585102 See also... Wielbark Goths were overwhelmingly of Scandinavian origin
Labels:
ancient ancestry,
ancient DNA,
Ancient Rome,
Central Europe,
Eastern Europe,
Goths,
Imperial Rome,
Medieval,
Middle Ages,
Migration Period,
Poland,
Polish,
Slavic,
Slavs,
Viking,
Viking Age,
Vikings,
Wielbark
Monday, February 12, 2024
The Nalchik surprise
If, like Iosif Lazaridis, you subscribe to the idea that the Yamnaya people carry early Anatolian farmer-related admixture that spread into Eastern Europe via the Caucasus, then I've got great news for you.
We now have a human sample from the Eneolithic site of Nalchik in the North Caucasus, labeled NL122, that packs well over a quarter of this type of ancestry (see here). Below is a quick G25/Vahaduo model to illustrate the point (please note that Turkey_N = early Anatolian farmers).
Target: Nalchik_Eneolithic:NL122 Distance: 2.1934% / 0.02193447 60.8 Russia_Steppe_Eneolithic 26.2 Turkey_N 13.0 Georgia_KotiasOn the other hand, if, again like Iosif Lazaridis, you subscribe to the idea that the Indo-European language spread into Eastern Europe via the Caucasus in association with this early Anatolian farmer-related admixture, then I've got terrible news for you. That's because NL122 is apparently dated to a whopping 5197-4850 BCE (see here). This dating might be somewhat bloated, possibly due to what's known as the reservoir effect, because the Nalchik archeological site is generally carbon dated to 4840–4820 BCE. However, even with the younger dating, this would still mean that early Anatolian farmer-related ancestry arrived in the North Caucasus, and thus in Eastern Europe, around 4,800 BCE at the latest. That's surprisingly early, and just too early to be relevant to any sort of Indo-European expansion from a necessarily even earlier Proto-Indo-Anatolian homeland somewhere south of the Caucasus. This means that NL122 effectively debunks Iosif Lazaridis' Indo-Anatolian hypothesis. Unless, that is, Iosif can provide evidence for a more convoluted scenario, in which there are at least two early Anatolian farmer-related expansions into Eastern Europe via the Caucasus, and the expansion relevant to the arrival of Indo-European speech came well after 5,000 BCE. I haven't done any detailed analyses of NL122 with formal stats and qpAdm. But my G25/Vahaduo runs suggest that it might be possible to model the ancestry of the Yamnaya people with around 10% admixture from a population similar to NL122.
Target: Russia_Samara_EBA_Yamnaya Distance: 3.4123% / 0.03412328 72.6 Russia_Progress_Eneolithic 18.2 Ukraine_N 9.2 Nalchik_EneolithicHowever, I don't subscribe to the idea that the Yamnaya people carry early Anatolian farmer-related admixture that spread into Eastern Europe via the Caucasus (on top of what is already found in Progress Eneolithic). Based on basic logic and a wide range of my own analyses, I believe that they acquired this type of ancestry from early European farmers, probably associated with the Trypillia culture. For instance...
Target: Russia_Samara_EBA_Yamnaya Distance: 3.2481% / 0.03248061 80.2 Russia_Progress_Eneolithic 13.6 Ukraine_Neolithic 6.2 Ukraine_VertebaCave_MLTrypillia 0.0 Nalchik_EneolithicAnother way to show this is with a Principal Component Analysis (PCA) that highlights a Yamnaya cline made up of the Yamnaya, Steppe Eneolithic and Ukraine Neolithic samples. As you can see, dear reader, there's no special relationship between the Yamnaya cline and Nalchik_Eneolithic. The Yamnaya samples, which are sitting near the eastern end of the Yamnaya cline, instead seem to show a subtle shift towards the Trypillian farmers. Indeed, I also don't exactly understand the recent infatuation among many academics, especially Iosif Lazaridis and his colleagues, with trying to put the Proto-Indo-Anatolian homeland somewhere south of the Caucasus. Considering all of the available multidisciplinary data, I'd say it still makes perfect sense to put it in the Sredny Stog culture of the North Pontic steppe, in what is now Ukraine. Please note that all of the G25 coordinates used in my models and the PCA are available HERE. See also... The Caucasus is a semipermeable barrier to gene flow
Saturday, January 13, 2024
Romans and Slavs in the Balkans (Olalde et al. 2023)
It's always amusing to see some random Jovan or Dimitar arguing online that Slavic speakers have been in the Balkans since at least the Neolithic.
Obviously, Slavic peoples only turned up in the Balkans during the early Middle Ages. It's just that their linguistic and genetic impact on the region was so profound that it may seem like they've been there forever.
A new paper at Cell by Olalde et al. makes this point well. See here.
That's not to say, however, that it's an ideal effort. The paper's qpAdm mixture models probably could've been more precise and realistic. Genes of the Ancients has a useful discussion on the topic here.
Interestingly, Olalde et al. admit that they can't detect much, if any, admixture from the Italian Peninsula in the Balkans, even in samples dating to the Roman period. And yet, this doesn't stop them from accepting that the Roman Empire had a massive cultural and demographic impact on the Balkans.
I also assume that, by extension, they don't deny that Latin was introduced into the Balkans from the Italian Peninsula.
That is, Latin spread into the Balkans without any noticeable genetic tracer dye, and it eventually gave rise to modern Romanian spoken by millions of people today in the eastern Balkans. This might be a useful data point to keep in mind when discussing the spread of Indo-European languages into Anatolia.
See also...
Dear Iosif, about that ~2%
Wednesday, December 20, 2023
Dear Harald #2
The ancIBD method paper from the David Reich Lab was just published in Nature (open access here). It's a very useful effort, but the authors are still somewhat confused about the origin of the Corded Ware culture (CWC) population. From the paper (emphasis is mine):
This direct evidence that most Corded Ware ancestry must have genealogical links to people associated with Yamnaya culture spanning on the order of at most a few hundred years is inconsistent with the hypothesis that the Steppe-like ancestry in the Corded Ware primarily reflects an origin in as-of-now unsampled cultures genetically similar to the Yamnaya but related to them only a millennium earlier.This is basically a straw man argument, because it's easy to debunk. So why put it in the paper? Well, as far as I can see, to make the idea that the CWC is derived from Yamnaya look more plausible. This idea, that the CWC is an offshoot of Yamnaya, seems to be the favorite explanation for the appearance of the CWC among the scientists at the David Reich Lab. However, I'd say they're facing a major problem with that, because the CWC and Yamnaya populations have largely different paternal origins. That is, CWC males mostly belong to Y-haplogroups R1a-M417 and R1b-L51, while Yamnaya males almost exclusively belong to Y-haplogroup R1b-Z2103. Indeed, as far as I know, there are no reliable instances of R1a-M417 or R1b-L51 in any published or yet to be published Yamnaya samples. But it is possible to reconcile the Y-haplogroup data with the ancIBD results if we assume that the peoples associated with the Corded Ware, Yamnaya and also Afanasievo cultures expanded from a genetically more diverse ancestral gene pool, each taking a specific subset of the variation with them. This gene pool would've existed somewhere in Eastern Europe, probably at the western end of the Pontic-Caspian steppe, at most a few hundred years before the appearance of the earliest CWC burials in what is now Poland. Moreover, the split between the CWC and Yamnaya populations need not have been a clean one, with long-range contacts and largely female-mediated mixing maintained for generations, adding to the already close genealogical links between them. Citation... Ringbauer, H., Huang, Y., Akbari, A. et al. Accurate detection of identity-by-descent segments in human ancient DNA. Nat Genet (2023). https://doi.org/10.1038/s41588-023-01582-w See also... On the origin of the Corded Ware people
Sunday, November 19, 2023
Musaeum Scythia on the Seima-Turbino Phenomenon
A few weeks ago bioRxiv published two preprints on the Seima-Turbino Phenomenon (see here and here).
I can't say much about these manuscripts until I see the relevant ancient DNA samples, and that might take some time.
However, for now, I will say that both preprints really need to emphasize the profound impact that the Sintashta-related early Indo-Iranian speakers had on the Seima-Turbino Phenomenon. This, of course, would require Wolfgang Haak and friends to pull their heads out of their behinds and admit that the proto-Indo-Iranian homeland was in Eastern Europe, not in Iran.
At the same time, it's likely that the Seima-Turbino Phenomenon originated deep in Siberia, and its inception was probably most closely associated with the West Siberian Hunter-Gatherer (WSHG) genetic component. It's important that the preprints emphasize this too.
Moreover, I can't see any convincing arguments in either preprint that the Seima-Turbino Phenomenon was mainly associated with proto-Uralic speakers, or even that it was an important vector for the spread of proto-Uralic. So there's not much point in forcing the Uralic angle on studies focused on the Seima-Turbino Phenomenon. Indeed, what we also need is an archaeogenetics paper dealing specifically with the proto-Uralic expansion.
Apart from that, I'd like to direct your attention to the fact that Musaeum Scythia has already written a fine blog post about these preprints:
Genomic insights into the Seima-Turbino Phenomenon
See also...
Finally, a proto-Uralic genome
The Uralic cline with kra001 - no projection this time
Slavs have little, if any, Scytho-Sarmatian ancestry
Friday, November 10, 2023
Wielbark Goths were overwhelmingly of Scandinavian origin
When used properly, Principal Component Analysis (PCA) is an extraordinarily powerful tool and one of the best ways to study fine-scale genetic substructures within Europe.
The PCA plot below is based on Global25 data and focuses on the genetic relationship between Wielbark Goths and Medieval Poles, including from the Viking Age, in the context of present-day European genetic variation.
I'd say that it's a wonderfully self-explanatory plot, but here are some key observations:
- the Wielbark Goths (Poland_Wielbark_IA) and Medieval Poles (Poland_Middle_Ages) are two distinct populations - moreover, the Wielbark Goths form a relatively compact Scandinavian-related cluster and must surely represent a homogenous population overwhelmingly of Scandinavian origin - on the other hand, the Medieval Poles form a more extensive and heterogeneous cluster that overlaps with present-day groups all the way from Central Europe to the East Baltic, and that's because they are likely to be in large part of mixed origin - I know for a fact that at least some of these early Poles harbor recent admixture, because their burials are similar to those of Vikings and their haplotypes have been shown to be partly of Scandinavian origin (see here) - one of the Wielbark females is an obvious genetic outlier (Poland_Wielbark_IA_outlier), and basically looks like a first generation mixture between a Goth and a Balt.Please note that the PCA is only based on relatively high quality genomes, so as not to confuse the picture with spurious results and noise. Also, all outliers with potentially significant ancestry from outside of Central, Eastern and Northern Europe were removed from the analysis. The relevant datasheet is available here. However, sanity checks are always important when studying complex topics like fine-scale genetic ancestry. To that end I've prepared a graph based on f3-statistics of the form f3(X,Cameroon_SMA,Estonia_BA)/(X,Cameroon_SMA,Ireland_Megalithic), that reproduces the key features of my PCA. The relevant datasheet is available here. Polish groups from the Middle Ages are marked with the MA suffix, while the Iron Age Wielbark Goths are marked with the IA suffix. If you're wondering why I plotted the f3-statistics that I did, take a look at this (all groups largely of Scandinavian origin are emboldened):
f3(X,Estonia_BA,Cameroon_SMA) Poland_Legowo_MA 0.226406 Poland_Ostrow_Lednicki_MA 0.225996 Poland_Plonsk_MA 0.225017 Poland_Trzciniec_Culture 0.224215 Poland_Lad_MA 0.224142 Poland_Viking 0.223838 Poland_Niemcza_MA 0.223659 Poland_Weklice_IA 0.223549 Poland_Kowalewko_IA 0.222584 Poland_Pruszcz_Gdanski_IA 0.222324 Sweden_Viking 0.222091 Russia_Viking 0.222042 Poland_Maslomecz_IA 0.221914 Norway_Viking 0.221825 Denmark_EarlyViking 0.221257 Denmark_Viking 0.221174 England_Viking 0.220979 f3(X,Ireland_Megalithic,Cameroon_SMA) Poland_Maslomecz_IA 0.219816 Poland_Weklice_IA 0.219501 Denmark_Viking 0.2192 Poland_Kowalewko_IA 0.219176 Poland_Ostrow_Lednicki_MA 0.218916 Norway_Viking 0.218854 Poland_Pruszcz_Gdanski_IA 0.218684 Sweden_Viking 0.218626 Denmark_EarlyViking 0.218529 England_Viking 0.218308 Russia_Viking 0.217999 Poland_Viking 0.217914 Poland_Plonsk_MA 0.217756 Poland_Lad_MA 0.217719 Poland_Legowo_MA 0.21765 Poland_Niemcza_MA 0.217001 Poland_Trzciniec_Culture 0.216551Interestingly, the Middle Bronze Age samples associated with the Trzciniec Culture (Poland_Trzciniec_Culture) show a closer genetic relationship to Medieval Poles than to Wielbark Goths or Northwestern Europeans. This is indeed the case both in terms of genome-wide and uniparental markers, including some very specific lineages under Y-chromosome haplogroup R1a. But that's a much more complex issue that I'll leave for another time. So please stay tuned. See also... Slavs have little, if any, Scytho-Sarmatian ancestry
Friday, September 22, 2023
The Caucasus is a semipermeable barrier to gene flow
The scientists at the David Reich Lab are a clever bunch. But they're not always on top of things, and this can be a problem.
For instance, they fail to understand that the Caucasus has effectively stymied human gene flow between Eastern Europe and West Asia through the ages. That is, the Caucasus is a semipermeable barrier to human gene flow.
Until they accept and understand this fact, they won't be able to accurately characterize the ancestry of the ancient human populations of the Pontic-Caspian (PC) steppe, including the Yamnaya people.
In turn, they also won't be able to locate the Indo-Anatolian homeland.
Now, the Caucasus isn't a barrier to gene flow because it's difficult to cross, and, indeed, many human populations have managed to cross it since the Upper Palaeolithic. As a result, the peoples of the North Caucasus are today genetically more similar to the populations of the Near East than Europe.
In fact, the clear genetic gap between most West Asian and Eastern European populations through the ages is actually caused by the extreme differentiation between the mountain ecology of the Caucasus and the steppe ecology of the PC steppe.
That is, the Caucasus is ecologically so different from the PC steppe that it has been practically impossible for human populations to make the transition from one to the other.
Indeed, it's important to understand that there's no reliable record of any prehistoric human population successfully making the transition from the mountain ecology to the steppe ecology in this part of the world.
In other words, contrary to claims by people like David Reich and David Anthony, there's no solid evidence of any significant prehistoric human migration from the Caucasus, or from south of the Caucasus, to Eastern Europe by hunter-gatherers, farmers or pastoralists.
But, you might ask, how on earth did the Yamnaya people get their significant Caucasus/Iranian-related admixture if not via a mass migration from the Caucasus and/or the Iranian Plateau, as is often argued by the above mentioned scholars and their many colleagues?
Well, obviously, the diffusion of alleles from one population to another can happen without migration. All that is needed is a contact zone between them.
The ancient DNA and archeological data currently available from the Caucasus and the PC steppe suggest to me that there was at least one such contact zone in this area bringing together the peoples of the mountain and steppe ecoregions. This allowed them to mix, probably gradually and over a long period of time, by and large without leaving their ecoregions.
Once the Caucasus alleles entered the steppe, they were spread around by local hunter-gatherers and pastoralists who were highly mobile and well adapted to the steppe ecology.
Someone should write a paper about this.
See also...
The Nalchik surprise
Understanding the Eneolithic steppe
Matters of geography
Labels:
ancient DNA,
Caucasus,
David Anthony,
David Reich,
David Reich Lab,
Eastern Europe,
ecology,
Indo-Anatolian,
Indo-European,
migration,
North Caucasus,
Pontic-Caspian steppe,
West Asia,
Yamnaya
Thursday, August 31, 2023
The story of the Khvalynsk people
I'm totally serious when I say that this video is more objective, informative and accurate than any peer-reviewed paper published to date when it comes to the genetic origins of the Khvalynsk people.
However, that's not to say it's perfect. I think it misses some important details. See here...
The Caucasus is a semipermeable barrier to gene flow
Dear David, Nick, Iosif...let's set the record straight
Understanding the Eneolithic steppe
Saturday, August 12, 2023
Indo-European crackpottery
I've now had the chance to read and digest the following two papers in Science about the origin of Indo-European languages:
Language trees with sampled ancestors support a hybrid model for the origin of Indo-European languages, Heggarty et al. The genetic history of the Southern Arc: A bridge between West Asia and Europe, Lazaridis, Alpaslan-Roodenberg et al.The Heggarty et al. paper is pure fluff. It offers nothing useful or even remotely interesting. For instance, the authors derive some Indo-European languages in Europe from Anatolian farmers and others from Caucasus hunter-gatherers (see here). This is not just exceedingly far fetched, but also obviously forced. Wolfgang Haak and Johannes Krause, you should be deeply ashamed of yourselves. I've already commented extensively about the Lazaridis, Alpaslan-Roodenberg et al. paper (for example, see here). But the one thing I need to add is that this paper is what it is due to the inherent bias of some of the lead authors to push the Indo-Anatolian homeland into West Asia. I won't even bother mentioning their names, because we all know who they are. See also... Crazy stuff Dear David, Nick, Iosif...let's set the record straight The story of R-V1636
Saturday, April 8, 2023
Dear Harald...
I've started analyzing the Identity-by-Descent (IBD) data from the recent Ringbauer et al. preprint (see here). Unfortunately, it'll take me a few weeks to do this properly, so I won't be able to write anything detailed on the topic for a while.
Meantime, this is the comment that I left for the authors at bioRxiv (at this time it's still being approved, but it should appear there within a day or so, possibly along with a reply from the authors):
Hello authors, Thanks for the interesting preprint and data. However, I'd like to see you address a couple of technical issues and perhaps one theoretical issue in the final manuscript: - the output you posted shows some unusual results, which are potentially false positives that appear to be concentrated among the shotgun and noUDG samples. I'm guessing that this is due to the same types of ancient DNA damage creating IBD-like patterns in these samples. If so, isn't there a risk that many or even most of the individuals in your analysis are affected by this problem to some degree, which might be skewing your estimates of genealogical relatedness between them? - many individuals from groups that have experienced founder effects, such as Ashkenazi Jews, appear to be close genetic cousins, even though they're not genealogical cousins. Basically, the reason for this is reduced haplotype diversity in such populations. Have you considered the possibility that at least some of the close relationships that you're seeing between individuals and populations might be exaggerated by founder effects? - thanks to ancient DNA we've learned that the Yamnaya phenomenon isn't just an archeological horizon, but also a closely related and genetically very similar group of people. Indeed, in my mind, ancient DNA has helped to redefine the Yamnaya concept, with Y-chromosome haplogroup R1b-Z2103 now being one of the key traits of the Yamnaya identity. So considering that the Corded Ware people are not rich in R1b-Z2103, and even the earliest Corded Ware individuals are somewhat different from the Yamnaya people in terms of genome-wide genetic structure, it doesn't seem right to keep claiming that the Corded Ware population is derived from Yamnaya. I can't see anything in your IBD data that would preclude the idea that the Corded Ware and Yamnaya peoples were different populations derived from the same as yet unsampled pre-Yamnaya/post-Sredny steppe group.See also... Dear Harald #2 On the origin of the Corded Ware people
Labels:
ancIBD,
ancient ancestry,
ancient DNA,
Corded Ware Culture,
CWC,
David Reich,
Eastern Europe,
haplotype,
Harald Ringbauer,
IBD,
Identity-by-Descent,
Nick Patterson,
Pontic-Caspian steppe,
Yamnaya
Monday, February 13, 2023
Dear David, Nick, Iosif...let me tell you about Yamnaya
Lazaridis, Alpaslan-Roodenberg et al. recently claimed that the Yamnaya people of the Pontic-Caspian (PC) steppe carried "substantial" ancestry from what is now Armenia or surrounds.
However, this claim is essentially false.
Only one individual associated with the Yamnaya culture shows an unambiguous signal of such ancestry. This is a female usually labeled Ukraine_Yamnaya_Ozera_o:I1917. The "o" suffix indicates that she is an outlier from the main Yamnaya genetic cluster.
Unlike I1917, typical Yamnaya individuals carry a few per cent of ancient European farmer admixture. This ancestry is only very distantly Armenian-related via Neolithic Anatolia (see here).
It's difficult for me to understand how Lazaridis, Alpaslan-Roodenberg et al. missed this. I suspect that they relied too heavily on formal statistics and overinterpreted their results.
Formal statistics are a very useful tool in ancient DNA work. Unfortunately, they're also a relatively blunt tool that often has problems distinguishing between similar sources of gene flow.
There are arguably better methods for studying fine scale ancestry, such as Principal Component Analysis (PCA).
Below is a somewhat special PCA featuring a wide range of ancient populations that plausibly might be relevant to the genetic origins of the Yamnaya people. Unlike most PCA with ancient samples, this PCA doesn't rely on any sort of projection, so that all of the actors are interacting with each other and directly affecting the outcome.
Here's another version of the same plot with a less complicated labeling system. Note that I designed this PCA specifically to differentiate between European populations and those from the Armenian highlands, the Iranian plateau and surrounds.
And here's a close up of the part of the plot that shows the Yamnaya cluster. This cluster is made up of samples associated with the Afanasievo, Catacomb, Poltavka and Yamnaya cultures. All of the individuals in this part of the plot are closely related, which is why they're so tightly packed together. The differentiation between them is caused by admixture from different groups mostly from outside of the PC steppe.
The Yamnaya cluster can be broadly characterized as a population that formed along the genetic continuum between the Eneolithic groups of the Progress region and Neolithic foragers from the Dnieper River valley (Progress_Eneolithic and Ukraine_N, respectively). However, this cluster also shows a slight western shift that is increasingly more pronounced in the Corded Ware samples. This shift is due to the aforementioned admixture from early European farmers.
Indeed, the plot reveals two parallel clines extending west from the Progress samples. One of the clines is made up of the Yamnaya cluster and the Corded Ware samples, and pulls towards the ancient European farmers. The other cline includes Ukraine_Yamnaya_Ozera_o:I1917 and pulls towards samples from the Armenian highlands and surrounds.
Being aware of these two clines and knowing how they came about is important to understanding the genetic prehistory of the PC steppe and indeed of much of Eurasia.
At some point, probably during the late Eneolithic, a Progress-related group experienced gene flow from the west and became the Yamnaya and Corded Ware populations. Sporadically, admixture from the Armenian highlands and the Iranian plateau also entered the PC steppe, giving rise to people like the Steppe Maykop outliers and Ukraine_Yamnaya_Ozera_o:I1917.
Unfortunately, this sort of PCA doesn't offer output suitable for mixture modeling, basically because the recent genetic drift shared by many of the samples creates significant noise.
However, to check that my inferences based on the plot are correct I can create composites with specific ancestry proportions to see how they behave. In the plot below Mix1 is 80% Progress_Eneolithic and 20% Iran_Hajji_Firuz_N, Mix2 is 80% Progress_Eneolithic and 20% Armenia_EBA_Kura_Araxes, while Mix3 is 80% Progress_Eneolithic, 15% Ukraine_N and 5% Hungary_MN_Vinca (Middle Neolithic farmers from the Carpathian Basin).
Obviously, we can't get Yamnaya by mixing Progress_Eneolithic with any ancients from the Armenian highlands or the Iranian plateau. On the other hand, Mix3 works quite well, at least in the first two dimensions. In some of the other dimensions genetic drift specific to Ukraine_N pulls it away from the Yamnaya cluster, but this is to be expected.
By the way, the plots were created with the excellent Vahaduo Custom PCA tool freely available here. It's well worth trying the interactive 3D option using my PCA data. The relevant datasheet is available here.
See also...
Dear David, Nick, Iosif...let's set the record straight
The Caucasus is a semipermeable barrier to gene flow
Sunday, November 13, 2022
A reappraisal of Ashkenazic maternal ancestry
Kevin Brook, who occasionally comments on this blog, has published a peer-reviewed book titled The Maternal Genetic Lineages of Ashkenazic Jews.
The book focuses on 129 mitochondrial (mtDNA) haplogroups that are found in present-day Ashkenazic Jews, and reveals that these lineages can be traced back to a wide range of places, such as Israel, Italy, Poland, Germany, North Africa, and China.
Ergo, it argues that both Israelites and converts to Judaism from a variety of gentile groups made lasting contributions to the Ashkenazic maternal gene pool. In Kevin's own words, the book also:
- shows that all Ashkenazim remain genetically linked to a significant degree to other types of Jewish populations, not only paternally but maternally as well - disproves the myth that Cossack rapists were responsible for any of the non-Israelite DNA in Ashkenazim - presents new DNA evidence in favor of a small contribution of Khazarian and Alan converts to Judaism to the Ashkenazic gene pool.That makes good sense based on what I've learned over the years from studying modern and ancient genome-wide Ashkenazic DNA. More information about Kevin's book is available at the Khazaria.com website HERE. See also... My take on the Erfurt Jews
Tuesday, November 1, 2022
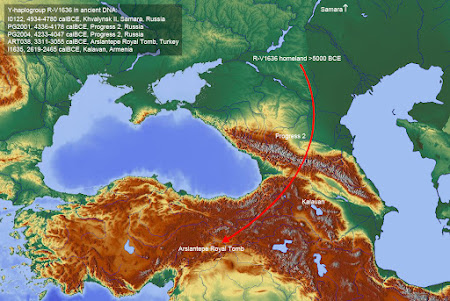
The story of R-V1636
Who wants to bet against this map? Keep in mind that ART038 (~3000 calBCE) remains the oldest sample with the V1636 and R1b Y-chromosome mutations in the West Asian ancient DNA record. Ergo, there's nothing to suggest that V1636 or R1b entered Eastern Europe from West Asia.
See also...
A tantalizing link
How relevant is Arslantepe to the PIE homeland debate?
Thursday, October 27, 2022
The Yassitepe challenge
This is about the only successful qpAdm model that I can find for the pair of Early Bronze Age (EBA) females from Yassitepe, Turkey, using a decent set of outgroups and markers. I wouldn't take it too literally, but it does suggest a potentially significant level of European ancestry, including some steppe ancestry, in these Yassitepe individuals.
TUR_Aegean_Yassitepe_EBA AZE_Caucasus_lowlands_LN 0.565±0.054 ROU_N 0.387±0.041 RUS_Progress_En 0.048±0.022 P-value 0.103248 Full outputIf anyone reading this can find a better, more convincing solution then I'd love to see it. Feel free to share it in the comments below. Obviously, both of the Yassitepe samples are from the recent Lazaridis, Alpaslan-Roodenberg et al. paper. Their EBA dating suggests that they might be relevant to the debate over the origins of Anatolian speakers, such as the Hittites and Luwians. See also... Dear Iosif, about that ~2% The precursor of the Trojans
Thursday, October 13, 2022
The Kura-Araxes people deserve better
When discussing the Kura-Araxes culture and its people it's important to understand these key points:
- there is Eastern European steppe ancestry in Kura-Araxes samples, and if you're not seeing it then you're not looking hard enough - Armenian Kura-Araxes samples are mainly a mixture between three different groups currently best represented in the ancient DNA record by ARM_Areni_C, IRN_Hajji_Firuz_C and RUS_Darkveti-Meshoko_En - ergo, most of the steppe ancestry in the Kura-Araxes population of what is now Armenia must have been mediated via local Chalcolithic groups like ARM_Areni_C - Kura-Araxes samples show Mesopotamian-related ancestry, and this mustn't be ignored.Oh, you don't believe it because you just read a big paper in Science claiming otherwise? Well, the authors of that paper, Lazaridis, Alpaslan-Roodenberg et al., used distal mixture models to study the ancestry of their Kura-Araxes samples, and such models can miss important details. Consider these three proximate mixture models for a relatively high quality and very homogenous Kura-Araxes sample set from the aforementioned paper. They were done with the qpAdm software
ARM_Kura-Araxes_Berkaber ARM_Areni_C 0.239±0.068 IRN_Hajji_Firuz_C 0.379±0.068 RUS_Darkveti-Meshoko_En 0.382±0.054 P-value 0.285122 (Pass) Full output ARM_Kura-Araxes_Berkaber IRN_Hajji_Firuz_C 0.569±0.051 RUS_Darkveti-Meshoko_En 0.363±0.058 RUS_Progress_En 0.068±0.020 P-value 0.20306 (Pass) Full output ARM_Kura-Araxes_Berkaber IRN_Hajji_Firuz_C 0.531±0.060 RUS_Darkveti-Meshoko_En 0.469±0.060 P-value 0.0132579 (Fail) Full outputSome caveats apply. For instance, the pass threshold (P-value ≥0.05) is arbitrary. But the point is that the models look much better with steppe-related and steppe reference populations (ARM_Areni_C and RUS_Progress_En, respectively). Moreover, the unique and vital Darkveti-Meshoko population is represented by just one individual. I also have the genotypes of his brother and sister, but relatives aren't allowed in these sorts of tests. Including a singleton in the analysis means that I can't use the inbreed: YES option, which apparently can be a bad thing. Nevertheless, these models do look very solid. Indeed, I can also model ARM_Kura-Araxes_Berkaber as practically 100% RUS_Maykop_Novosvobodnaya, perhaps with some excess ARM_Areni_C-related input.
ARM_Kura-Araxes_Berkaber ARM_Areni_C 0.094±0.087 RUS_Maykop_Novosvobodnaya 0.906±0.087 P-value 0.284259 (Pass) Full ouputThis makes good sense, because RUS_Maykop_Novosvobodnaya can also be modeled solidly as a mixture between IRN_Hajji_Firuz_C, RUS_Darkveti-Meshoko_En and RUS_Progress_En.
RUS_Maykop_Novosvobodnaya IRN_Hajji_Firuz_C 0.614±0.056 RUS_Darkveti-Meshoko_En 0.307±0.064 RUS_Progress_En 0.080±0.022 P-value 0.141468 (Pass) Full outputI don't know whether the genetic relationship between ARM_Kura-Araxes_Berkaber and RUS_Maykop_Novosvobodnaya shown in my model is due to Maykop ancestry in the former. It might just be a coincidence in the sense that the same or similar processes led to the formation of both groups. Feel free to let me know your thoughts about that in the comments. The fact that the Kura-Araxes people harbored steppe ancestry might be very important in the debate over the location of the so called Indo-Anatolian homeland. For instance, it's possible that the proto-Anatolian language spread from the North Caucasus into Anatolia via the Kura-Araxes culture. But, admittedly, such a solution doesn't have strong support from historical linguistics data, which suggest that the Indo-Anatolian homeland was located in what is now Ukraine and that Anatolian speakers entered West Asia via the Balkans:
Indo-European cereal terminology suggests a Northwest Pontic homeland for the core Indo-European languagesSee also... R-V1636: Eneolithic steppe > Kura-Araxes? Dear Iosif...Yamnaya But Iosif, what about the Phrygians?
Thursday, October 6, 2022
Balto-Slavs and Sarmatians in the Battle of Himera
G25 coordinates for most of the samples from the recent Reitsema et al. paper are available in a text file here. They're also in the G25 datasheets at the usual link here.
A basic distance analysis with the G25 data at Vahaduo shows that the two samples labeled Himera_480BCE_3 are either early Balts or Slavs. I suspect that they're Slavs, because I believe that early Slavs had this type of Baltic-like genetic structure before mixing with their non-Slavic-speaking neighbors. Well, that's my pet theory for now, so take it or leave it.
Distance to: ITA_Sicily_Himera_480BCE_3:I10943 0.03393838 HUN_IA_La_Tene_o:I18226 0.03572886 DEU_MA_Krakauer_Berg:KRA001 0.03618075 RUS_Pskov_VA:VK159 0.03899963 SWE_Gotland_VA:VK463 0.03915018 Baltic_EST_IA:s19_V12_1 Distance to: ITA_Sicily_Himera_480BCE_3:I10949 0.03573636 HUN_IA_La_Tene_o3:I25524 0.03698768 HUN_IA_La_Tene_o:I18226 0.03732752 SWE_Skara_VA:VK397 0.03767022 Baltic_EST_IA:s19_V12_1 0.03772687 DEU_MA_Krakauer_Berg:KRA001On the other hand, I'm almost certain that the two Himera_480BCE_4 samples are Sarmatians. The good old G25 does it again!
Distance to: ITA_Sicily_Himera_480BCE_4:I10944 0.03100861 KAZ_Segizsay_Sarmatian:SGZ002 0.03548059 MDA_Sarmatian:I11925 0.03619219 RUS_Urals_Sarmatian:MJ56 0.03626538 RUS_Urals_Sarmatian:chy001 0.03904260 RUS_Urals_Sarmatian:MJ41 Distance to: ITA_Sicily_Himera_480BCE_4:I10947 0.02989458 RUS_Urals_Sarmatian:MJ43 0.03052790 RUS_Urals_Sarmatian:chy002 0.03170622 KAZ_Kangju:DA226 0.03288789 TUR_BlackSea_Samsun_Anc_C:I4529 0.03310149 KAZ_Aigyrly_Sarmatian:AIG003See also... Slavic-like Medieval Germans
Labels:
ancient ancestry,
ancient DNA,
Ancient Greece,
Balto-Slavic,
battle,
East Baltic,
Eastern Europe,
Eastern European steppe,
Himera,
Italy,
Mediterranean,
Northeastern Europe,
Sarmatian,
Sicily,
Slavic,
Slavs
Monday, September 19, 2022
Dear Iosif...Yamnaya
Even though the Yamnaya culture probably originated in what is now Ukraine, the earliest Yamnaya samples currently available are from the modern-day Samara region of Russia. They mostly date to around 3,000 BCE. I can analyze their ancestry using Principal Component Analysis (PCA) data.
Target: RUS_Yamnaya_Samara Distance: 3.2816% / 0.03281581 81.0 RUS_Progress_En 14.4 UKR_N 4.6 HUN_Vinca_MN 0.0 ARM_Aknashen_N 0.0 ARM_Masis_Blur_N 0.0 AZE_Caucasus_lowlands_LN 0.0 BGR_C 0.0 BGR_Dzhulyunitsa_N 0.0 IRN_Ganj_Dareh_N 0.0 IRN_Hajji_Firuz_C 0.0 IRN_Seh_Gabi_C 0.0 IRN_Tepe_Abdul_Hosein_N 0.0 IRN_Wezmeh_N 0.0 RUS_Darkveti-Meshoko_En 0.0 RUS_Maykop 0.0 RUS_Maykop_Late 0.0 RUS_Maykop_NovosvobodnayaThe above results show exactly zero ancestry from West Asia. Admittedly, both RUS_Progress_En and HUN_Vinca_MN are European ancients with significant West Asian-related ancestry. However, this ancestry is very distantly West Asian-related, and, for instance, it almost certainly has no relevance to the Indo-Anatolian homeland debate. The Afanasievo culture of Central Asia is regarded to have been an early offshoot of the Yamnaya culture. A good number of Afanasievo samples are available, so let's have a look if their results match those of the Yamnaya folks. And indeed they do, since BGR_C is very similar to HUN_Vinca_MN.
Target: RUS_Afanasievo Distance: 3.4055% / 0.03405499 84.0 RUS_Progress_En 11.4 UKR_N 4.6 BGR_C 0.0 ARM_Aknashen_N 0.0 ARM_Masis_Blur_N 0.0 AZE_Caucasus_lowlands_LN 0.0 BGR_Dzhulyunitsa_N 0.0 HUN_Vinca_MN 0.0 IRN_Ganj_Dareh_N 0.0 IRN_Hajji_Firuz_C 0.0 IRN_Seh_Gabi_C 0.0 IRN_Tepe_Abdul_Hosein_N 0.0 IRN_Wezmeh_N 0.0 RUS_Darkveti-Meshoko_En 0.0 RUS_Maykop 0.0 RUS_Maykop_Late 0.0 RUS_Maykop_NovosvobodnayaTo try this at home, stick the PCA data in the text file here into the relevant fields here and cranck up the "Cycles" to 4X. You should see exactly zero ancestry from West Asia every time. I can, more or less, reproduce these results with tools that are routinely used in peer reviewed papers. Below is a table of mixture models produced with the qpAdm software. I set the pass threshold to P ≥0.05, which is an arbitrary value, but the pattern is clear. The full output from each qpAdm run is available here. Importantly, qpAdm needs to be fed the relevant "right pop" outgroups to be able to discriminate accurately between reference populations.
right pops: CMR_Shum_Laka_8000BP MAR_Taforalt Levant_Natufian IRN_Ganj_Dareh_N Levant_PPNB TUR_Marmara_Barcin_N HUN_Starcevo_N HUN_Koros_N SRB_Iron_Gates_HG Iberia_Southeast_Meso RUS_Karelia_HG RUS_West_Siberia_HG RUS_Boisman_MN MNG_North_N TWN_Hanben BRA_LapaDoSanto_9600BPSo, for instance, if one were to use in this role the modern-day Mbuti people, as opposed to, say, the ancient hunter-gatherers of Shum Laka, one might find that many models look statistically better than they should. And then one might also find that the Yamnaya samples carry significant West Asian ancestry. Actually, I'm not opposed to the idea of some West Asian ancestry in Yamnaya. Indeed, considering the extraordinary mobility of the Yamnaya people and their Eneolithic predecessors on the Pontic-Caspian steppe, it would be unusual if they didn't come into close contact and mix, to some degree, with their neighbors from West Asia. However, based on everything I've seen, from uniparental markers to different types of autosomal genetic tests, it's clear to me that there's no substantial West Asian ancestry in any Yamnaya samples, except for an outlier female from modern-day Ozera, Ukraine (see here). Admittedly, ancient DNA does have a habit of throwing curveballs, so I'm eagerly awaiting new Eneolithic samples from the Pontic-Caspian steppe, particularly those associated with the Yamnaya-like Sredni Stog culture, to help finally settle this issue. Believe it or not, a contact recently sent me a supposedly unpublished female sample from a ~4,200 BCE Sredni Stog burial in modern-day Igren, east central Ukraine. So what the hell, let's assume for the time being that this sample is genuine. This is how Miss Sredni Stog behaves in my PCA mixture test.
Target: UKR_Sredni_Stog Distance: 4.0769% / 0.04076877 75.6 RUS_Progress_En 17.8 UKR_N 6.6 HUN_Vinca_MN 0.0 ARM_Aknashen_N 0.0 ARM_Masis_Blur_N 0.0 AZE_Caucasus_lowlands_LN 0.0 BGR_C 0.0 BGR_Dzhulyunitsa_N 0.0 HUN_Vinca_MN 0.0 IRN_Ganj_Dareh_N 0.0 IRN_Hajji_Firuz_C 0.0 IRN_Seh_Gabi_C 0.0 IRN_Tepe_Abdul_Hosein_N 0.0 IRN_Wezmeh_N 0.0 RUS_Darkveti-Meshoko_En 0.0 RUS_Maykop 0.0 RUS_Maykop_Late 0.0 RUS_Maykop_NovosvobodnayaWow, just wow. Have we actually found Miss Proto-Yamnaya? What does qpAdm have to say in the matter?
UKR_Sredni_Stog HUN_Vinca_MN 0.034±0.028 RUS_Progress_En 0.796±0.045 UKR_N 0.170±0.034 P-value 0.41088Again, this is an excellent match with the results from my PCA test, especially if we take into account the standard errors. However, with qpAdm it's also possible to model this individual's ancestry as part West Asian.
UKR_Sredni_Stog AZE_Caucasus_lowlands_LN 0.056±0.039 RUS_Progress_En 0.761±0.061 UKR_N 0.183±0.036 P-value 0.465667As I pointed out above, it's plausible for such people to harbor some West Asian ancestry, but I'm very sceptical that this is really the case here, despite the rather solid qpAdm statistical fit. That's because UKR_Sredni_Stog is not a high quality sample, and, from my experience, qpAdm often has problems analyzing fine scale ancestry in singletons or even small groups that show excess DNA damage and/or offer much less than a million markers. See also... Dear Iosif, about that ~2% But Iosif, what about the Phrygians?
Sunday, September 4, 2022
But Iosif, what about the Phrygians?
A paper in Science co-authored by around 200 scientists from some of the world's top academic institutions surely must mean something, right? Not necessarily.
In this short blog post I'll try to explain, as simply as I can, why the Lazaridis, Alpaslan-Roodenberg et al. paper doesn't get us any closer to solving the riddle of the so called Indo-Anatolian homeland.
However, it must be said that the paper does include many interesting and valuable samples. I'll be using six of these samples, labeled TUR_C_Gordion_Anc, to argue my case.
The TUR_C_Gordion_Anc sample set is from Gordion, the capital of ancient Phrygia, and thus, in all likeliness, it represents Phrygian speakers.
Phrygian is an Indo-European language and the leading hypothesis is that it originated in the Balkans.
In terms of fine scale ancestry, TUR_C_Gordion_Anc can be reliably divided into two genetic clusters. In the Principal Component Analysis (PCA) below these clusters are labeled TUR_C_Gordion_Anc1 and TUR_C_Gordion_Anc2.
Note that TUR_C_Gordion_Anc1 is obviously pulling away from TUR_C_Gordion_Anc2 towards samples from the Balkans. I've used ancient samples from what is now North Macedonia, labeled MKD_Anc, to represent the Balkans. To see an interactive version of the plot, paste the PCA coordinates from here into the relevant field here.
Visually, this is not an especially dramatic outcome, but it's an incredible result nonetheless, because it shows that even a few ancient samples can help to solve an age old mystery.
Across many dimensions of genetic variation, the shift in the PCA from TUR_C_Gordion_Anc1 to TUR_C_Gordion_Anc2 represents about 20% admixture from the Balkans, and about 8% from the Eastern European steppe. That's plenty enough to corroborate the linguistic hypothesis that the Phrygians originated in the Balkans, and that some of their ancestors came from the steppe. The mixture models below were done with the tools here.
Target: TUR_C_Gordion_Anc1 Distance: 1.6634% / 0.01663373 40.6 Kura-Araxes_ARM_Kaps 22.2 Anatolia_Barcin_N 21.8 MKD_Anc 13.6 Levant_PPNB 1.4 IRN_Ganj_Dareh_N 0.4 Han Target: TUR_C_Gordion_Anc1 Distance: 1.7109% / 0.01710904 40.2 Kura-Araxes_ARM_Kaps 37.8 Anatolia_Barcin_N 12.4 Levant_PPNB 8.0 Yamnaya_RUS_Samara 1.2 IRN_Ganj_Dareh_N 0.4 Han Target: TUR_C_Gordion_Anc2 Distance: 2.0293% / 0.02029339 51.0 Kura-Araxes_ARM_Kaps 26.8 Anatolia_Tepecik_Ciftlik_N 17.6 Anatolia_Barcin_N 4.6 Levant_PPNBSurprisingly, Lazaridis, Alpaslan-Roodenberg et al. didn't have much to say about this topic. This quote basically sums it up:
However, in individuals from Gordion, a Central Anatolian city that was under the control of Hittites before becoming the Phrygian capital and then coming under the control of Persian and Hellenistic rulers, the proportion of Eastern hunter-gatherer ancestry is only ~2%, a tiny fraction for a region controlled by at least four different Indo-European–speaking groups.I have no doubt that Lazaridis, Alpaslan-Roodenberg et al. can run a very decent PCA, and then blow it up to a size big enough to show that the Gordion samples represent two genetically somewhat distinct groups. I'm also sure that, if they really try, they can locate significant levels of proximate and relevant European ancestry in some of these samples. They don't have to use my methods; they can use any methods they like. My point is that they won't find much if they're just looking for genetic signals from the Upper Paleolithic or Mesolithic. Now, considering the way that the Phrygian question was treated by Lazaridis, Alpaslan-Roodenberg et al., despite the fact that they managed to sequence a few likely Phrygian speakers from none other than the Phrygian capital, let's not pretend that their paper brought us any closer to understanding the genetic origins of Anatolian speakers or pinpointing their ancestral homeland. In order to even try to solve these problems with ancient DNA, we need a wide range of samples from Hittite, Luwian and other key sites where Anatolian languages were spoken. And then we must analyze them properly. I'm guessing that Lazaridis, Alpaslan-Roodenberg et al. went out of their way to get such samples, but for one reason or another they failed. If so, that's OK, but I have a feeling that even if they got them, they wouldn't know what to do with them, because at best these samples would only show ~2% Eastern hunter-gatherer ancestry. Haha. For what it's worth, I believe that the ancient data in the Lazaridis, Alpaslan-Roodenberg et al. paper point to the North Pontic steppe as the Indo-Anatolian homeland, and I'll lay out my arguments in an upcoming blog post. See also... Dear Iosif...Yamnaya Dear Iosif, about that ~2% Dear Iosif... Dear Iosif #2 Dear Iosif #3
Subscribe to:
Posts (Atom)