search this blog
Showing posts with label Corded Ware. Show all posts
Showing posts with label Corded Ware. Show all posts
Monday, February 13, 2023
Dear David, Nick, Iosif...let me tell you about Yamnaya
Lazaridis, Alpaslan-Roodenberg et al. recently claimed that the Yamnaya people of the Pontic-Caspian (PC) steppe carried "substantial" ancestry from what is now Armenia or surrounds.
However, this claim is essentially false.
Only one individual associated with the Yamnaya culture shows an unambiguous signal of such ancestry. This is a female usually labeled Ukraine_Yamnaya_Ozera_o:I1917. The "o" suffix indicates that she is an outlier from the main Yamnaya genetic cluster.
Unlike I1917, typical Yamnaya individuals carry a few per cent of ancient European farmer admixture. This ancestry is only very distantly Armenian-related via Neolithic Anatolia (see here).
It's difficult for me to understand how Lazaridis, Alpaslan-Roodenberg et al. missed this. I suspect that they relied too heavily on formal statistics and overinterpreted their results.
Formal statistics are a very useful tool in ancient DNA work. Unfortunately, they're also a relatively blunt tool that often has problems distinguishing between similar sources of gene flow.
There are arguably better methods for studying fine scale ancestry, such as Principal Component Analysis (PCA).
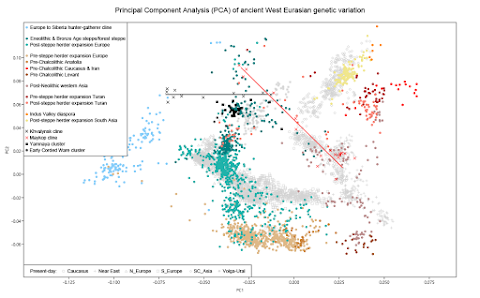
Below is a somewhat special PCA featuring a wide range of ancient populations that plausibly might be relevant to the genetic origins of the Yamnaya people. Unlike most PCA with ancient samples, this PCA doesn't rely on any sort of projection, so that all of the actors are interacting with each other and directly affecting the outcome.
Here's another version of the same plot with a less complicated labeling system. Note that I designed this PCA specifically to differentiate between European populations and those from the Armenian highlands, the Iranian plateau and surrounds.
And here's a close up of the part of the plot that shows the Yamnaya cluster. This cluster is made up of samples associated with the Afanasievo, Catacomb, Poltavka and Yamnaya cultures. All of the individuals in this part of the plot are closely related, which is why they're so tightly packed together. The differentiation between them is caused by admixture from different groups mostly from outside of the PC steppe.
The Yamnaya cluster can be broadly characterized as a population that formed along the genetic continuum between the Eneolithic groups of the Progress region and Neolithic foragers from the Dnieper River valley (Progress_Eneolithic and Ukraine_N, respectively). However, this cluster also shows a slight western shift that is increasingly more pronounced in the Corded Ware samples. This shift is due to the aforementioned admixture from early European farmers.
Indeed, the plot reveals two parallel clines extending west from the Progress samples. One of the clines is made up of the Yamnaya cluster and the Corded Ware samples, and pulls towards the ancient European farmers. The other cline includes Ukraine_Yamnaya_Ozera_o:I1917 and pulls towards samples from the Armenian highlands and surrounds.
Being aware of these two clines and knowing how they came about is important to understanding the genetic prehistory of the PC steppe and indeed of much of Eurasia.
At some point, probably during the late Eneolithic, a Progress-related group experienced gene flow from the west and became the Yamnaya and Corded Ware populations. Sporadically, admixture from the Armenian highlands and the Iranian plateau also entered the PC steppe, giving rise to people like the Steppe Maykop outliers and Ukraine_Yamnaya_Ozera_o:I1917.
Unfortunately, this sort of PCA doesn't offer output suitable for mixture modeling, basically because the recent genetic drift shared by many of the samples creates significant noise.
However, to check that my inferences based on the plot are correct I can create composites with specific ancestry proportions to see how they behave. In the plot below Mix1 is 80% Progress_Eneolithic and 20% Iran_Hajji_Firuz_N, Mix2 is 80% Progress_Eneolithic and 20% Armenia_EBA_Kura_Araxes, while Mix3 is 80% Progress_Eneolithic, 15% Ukraine_N and 5% Hungary_MN_Vinca (Middle Neolithic farmers from the Carpathian Basin).
Obviously, we can't get Yamnaya by mixing Progress_Eneolithic with any ancients from the Armenian highlands or the Iranian plateau. On the other hand, Mix3 works quite well, at least in the first two dimensions. In some of the other dimensions genetic drift specific to Ukraine_N pulls it away from the Yamnaya cluster, but this is to be expected.
By the way, the plots were created with the excellent Vahaduo Custom PCA tool freely available here. It's well worth trying the interactive 3D option using my PCA data. The relevant datasheet is available here.
See also...
Dear David, Nick, Iosif...let's set the record straight
The Caucasus is a semipermeable barrier to gene flow
Wednesday, February 2, 2022
The PIE homeland controversy: February 2022 status report
I think we'll see the emergence of two main competing proto-Indo-European (PIE) homeland theories over the next few years:
- a homeland in the Eneolithic North Caucasus, and the spread of Anatolian languages into West Asia with Maykop-related ancestry - a homeland in the North Pontic region, possibly within the Eneolithic Sredny Stog archeological culture, and the spread of Anatolian languages into West Asia via the Balkans.Both theories have support from ancient DNA. Some of it has already been published (for instance, see here). At this point, I can see myself firmly in the North Pontic camp, even if it turns out that North Pontic-related ancestry only made a fleeting impact on Bronze Age Anatolia. After all, there's no direct relationship between genes and languages, so to prove that Anatolian languages came from the North Pontic, there's no need for North Pontic-related ancestry to persist in Anatolia, as long as we have solid evidence that people with this type of ancestry moved there at the right time. In my mind, for now, the Maykop culture provides an excellent explanation for non-Indo-European influences in PIE, and there's no need to make it Indo-European speaking, let alone PIE speaking. See also... The PIE homeland controversy: June 2021 status report
Friday, January 21, 2022
Yamnaya is from Europe, but it's really from Asia
I was about to post a comment under a new preprint at bioRxiv, but the comment section isn't there anymore. Hopefully, this is just a temporary glitch.
The preprint in question is titled Reconstructing the spatiotemporal patterns of admixture during the European Holocene using a novel genomic dating method [LINK]. It's co-authored by Harvard/Broad MIT scientist Nick Patterson who occasionally comments at this blog.
My impression is that the authors see the people associated with the Yamnaya culture as Asians who simply used "far" Eastern Europe as a springboard to expand into other parts of Europe.
If so, they're dead wrong.
There are at least three arguments why the Yamnaya population should be seen as quintessentially European:
- its home was initially and overwhelmingly the Pontic-Caspian steppe, which is entirely located within the present-day borders of Europe - Yamnaya genomes are clearly different from those of older populations native to nearby parts of Asia, and, in fact, these differences show a very strong correlation with the present-day borders between Europe and Asia - the Yamnaya people weren't a new population in Europe by any stretch, but must have been overwhelmingly derived from the very similar Eneolithic peoples of the Pontic-Caspian steppe and/or the nearby forest steppe, both of which are located in Eastern Europe.And yet, this is what the preprint claims:
The beginning of the Bronze Age was a period of major cultural and demographic change in Eurasia, accompanied by the spread of Yamnaya Steppe Pastoralist-related ancestry from Pontic-Caspian steppes into Europe and South Asia (16).In fact, what really happened at this time was that Yamnaya steppe pastoralist-related ancestry spread from Eastern Europe to other parts of Europe, as well as to Central and West Asia. The preprint does eventually explain that present-day South Asians derive their Yamnaya-related ancestry from a later eastward expansion of the European Corded Ware culture (CWC), but it completely ignores the fact that the Afanasievo culture was the result of the initial eastward expansion from Europe to Asia. That is, the ancestors of the Afanasievo people were recent migrants from the Pontic-Caspian steppe to Central Asia and Siberia. There's also this:
Over the following millennium, the Yamnaya-derived groups of the Corded Ware Complex (CWC) and Bell Beaker complex (BBC) cultures brought Steppe pastoralist-related ancestry to Europe.Seriously? Both the CWC and BBC, just like the Yamnaya culture, were from Europe. In fact, as per above, the descendants of the CWC expanded into Asia. And this:
The second major migration occurred when populations associated with the Yamnaya culture in the Pontic-Caspian steppe expanded to central and western Europe from far eastern Europe.The authors basically admit here that Yamnaya came from Eastern Europe, but they call it "far" Eastern Europe. Perhaps they know something I don't, but as things stand, there's no evidence that Yamnaya came from "far" Eastern Europe. In fact, the emerging consensus based on ancient DNA, including pre-publication data, is that Yamnaya may have originated in what is now Ukraine. In my opinion, Ukraine isn't located in "far" Eastern Europe, but more or less in the middle of it. Inexplicably, this is what they say about the genetic origins of the Yamnaya and Afanasievo peoples:
These groups were likely the result of a genetic admixture between the descendants of EHG-related groups and CHG-related groups associated with the first farmers from Iran (8, 22, 36). ... Thus, we combined all early Steppe pastoralist individuals in one group to obtain a more precise estimate for the genetic formation of proto-Yamnaya of ~4,400 to 4,000 BCE (Figure 2). These dates are noteworthy as they pre-date the archeological evidence by more than a millennium (37) and have important implications for understanding the origin of proto-Pontic Caspian cultures and their spread to Europe and South Asia.Not really. Like I said, the Yamnaya population was overwhelmingly derived from the Eneolithic peoples of the Eastern European steppe and/or forest steppe. And these Yamnaya-like Eneolithic peoples were spread out across a vast area of Eastern Europe by at least ~4,500 BCE. Some of their genomes have been available for several years, and many more are on the way. It is possible that the Yamnaya and Afanasievo genotype formed in 4,400-4,000 BCE, but if so, then this was due to mixing between the Eneolithic steppe peoples and nearby European farmers. That's because the difference between the Yamnaya and Eneolithic steppe genotypes is minor (~15%) European farmer admixture in the former. The really interesting puzzle is exactly where and when the peculiar Eneolithic steppe genotype came into being. Any ideas Dr Patterson? See also... Matters of geography Understanding the Eneolithic steppe
Labels:
Afanasevo,
Afanasievo,
ancient DNA,
Corded Ware,
Corded Ware Culture,
CWC,
Eastern Europe,
Eurasia,
Eurasian steppe,
Indo-European,
Pontic-Caspian steppe,
Proto-Indo-European,
Yamna,
Yamnaya
Friday, August 27, 2021
R1a vs R1b in third millennium BCE Central Europe (Papac et al. 2021)
R1a-M417 and R1b-L51 are by far the most important Y-chromosome haplogroups in Europe today. More precisely, R1a-M417 dominates in Eastern Europe, while R1b-L51 in Western Europe.
It's been obvious for a while now, at least to me, that both of these Y-haplogroups are closely associated with the men of the Late Neolithic Corded Ware culture (CWC). Indeed, in my mind they're the main genetic signals of its massive expansion, probably from a homeland somewhere north of the Black Sea in what is now Ukraine.
I'm still not exactly sure how the east/west dichotomy between R1a and R1b emerged in Europe, but, thanks to a new paper by Papac et al. at Science Advances, at least now I have a working hypothesis about that. Below is a quote from the said paper, emphasis is mine:
In addition to autosomal genetic changes through time, we observe a sharp reduction in Y-chromosomal diversity going from five different lineages in early CW to a dominant (single) lineage in late CW (Fig. 4A). We used forward simulations to explore the demographic scenarios that could account for the observed reduction in Y-chromosomal diversity. Performing 1 million simulations of a population with a starting frequency of R1a-M417(xZ645) centered around the observed starting frequency in Bohemia_CW_Early (3 of 11, 0.27), we assessed the plausibility of this lineage reaching the observed frequency in Bohemia_CW_Late (10 of 11, 0.91) in the time frame of 500 years under a model of a closed population and random mating (Materials and Methods). We reject the “neutral” hypothesis, i.e., that this change in frequency occurred by chance, given a wide range of plausible population sizes. Instead, our results suggest that R1a-M417(xZ645) was subject to a nonrandom increase in frequency, resulting in these males having 15.79% (4.12 to 44.42%) more surviving offspring per generation relative to males of other Y-haplogroups. We also find that this change in Y chromosome frequency is extreme compared to the changes in allele frequencies at fully covered autosomal 1240k sites within the same males, suggesting a process that disproportionately affected Y-chromosomal compared to autosomal genetic diversity, ruling out a population bottleneck as the likely cause. Our results suggest that the Y-lineage diversity in early CW males was supplanted by a nonrandom process [selection, social structure, or influx of nonlocal R1a-M417(xZ645) lineages] that drove the collapse in Y-chromosomal diversity. A simultaneous decline of Y-chromosomal diversity dating to the Neolithic has been observed across most extant Y-haplogroups (64), possibly due to increased conflict between male-mediated patrilines (65). We view that changes in social structure (e.g., an isolated mating network with strictly exclusive social norms) could be an alternative cause but would be difficult to distinguish in the underlying model parameters.Right, so even though the CWC was clearly a community of closely related groups, there must have been some competition between its different clans. And since these clans were highly patriarchal and patrilineal, this competition probably led to different paternal lineages dominating different parts of the CWC horizon, with M417 becoming especially common in the east and L51 in the west. Of course, the expansions of post-Corded Ware groups, such as the M417-rich Slavs in Eastern Europe and L51-rich Celts in Western Europe, were also instrumental in creating Europe's R1a/R1b dichotomy, but obviously these groups were in large part the heirs of the CWC. By the way, most of the samples from Papac et al. are already in the Global25 datasheets linked here. Look for the labels listed here. Below is a plot made from the Global25 data courtesy of regular commentator Matt. Citation: L. Papac, M. Ernée, M. Dobeš, M. Langová, A. B. Rohrlach, F. Aron, G. U. Neumann, M. A. Spyrou, N. Rohland, P. Velemínský, M. Kuna, H. Brzobohatá, B. Culleton, D. Daněček, A. Danielisová, M. Dobisíková, J. Hložek, D. J. Kennett, J. Klementová, M. Kostka, P. Krištuf, M. Kuchařík, J. K. Hlavová, P. Limburský, D. Malyková, L. Mattiello, M. Pecinovská, K. Petriščáková, E. Průchová, P. Stránská, L. Smejtek, J. Špaček, R. Šumberová, O. Švejcar, M. Trefný, M. Vávra, J. Kolář, V. Heyd, J. Krause, R. Pinhasi, D. Reich, S. Schiffels, W. Haak, Dynamic changes in genomic and social structures in third millennium BCE central Europe. Sci. Adv. 7, eabi6941 (2021). See also... On the origin of the Corded Ware people Understanding the Eneolithic steppe Conan the Barbarian probably belonged to Y-haplogroup R1a
Labels:
ancient DNA,
Balto-Slavic,
Celtic,
Central Europe,
Corded Ware,
Corded Ware Culture,
CWC,
Eastern Europe,
Germanic,
Indo-European,
Northern Europe,
Proto-Indo-European,
R1a-M417,
R1b-L151,
R1b-L51,
Slavic,
Slavs
Tuesday, July 20, 2021
On the origin of the Corded Ware people
There's been a lot of talk lately about the finding that the peoples associated with the Corded Ware and Yamnaya archeological cultures were genetic cousins (for instance, see here). As I've already pointed out, this is an interesting discovery, but, at this stage, it's difficult to know what it means exactly.
It might mean that the Yamnayans were the direct predecessors of the Corded Ware people. Or it might just mean that, at some point, the Corded Ware and Yamnaya populations swapped women regularly (that is, they practiced female exogamy with each other).
In any case, I feel that several important facts aren't being taken into account by most of the interested parties. These facts include, in no particular order:
- despite being closely related, the Corded Ware and Yamnaya peoples were highly adapted to very different ecological zones - temperate forests and arid steppes, respectively - and this is surely not something that happened within a few years and probably not even within a couple of generations - both the Corded Ware and Yamnaya populations expanded widely and rapidly at around the same time, but never got in each others way, probably because they occupied very different ecological niches - despite sharing the R1b Y-chromosome haplogroup, their paternal origins were quite different, with Corded Ware males rich in R1a-M417 and R1b-L51 and Yamnaya males rich in R1b-Z2103 and I2a-L699I suppose it's possible that the Corded Ware people were overwhelmingly and directly derived from the Yamnaya population. But right now my view is that, even if they were, then the Yamnaya population that they came from was quite different from the classic, R1b-Z2103-rich Yamnaya that spread rapidly across the steppes. Indeed, perhaps what we're dealing with here is a very early (proto?) Yamnaya gene pool located somewhere in the border zone between the forests and the steppes, that then split into two main sub-populations, with one of these groups heading north and the other south? I do wonder what David Anthony would say if he was made aware of the above mentioned facts? Then again, perhaps he's already aware of them, and simply chose to ignore them when formulating his latest theory about the origin of the Corded Ware people? See also...
Monday, June 28, 2021
The PIE homeland controversy: June 2021 status report
Archeologist David Anthony has made several appearances online recently to promote his theories about the origins of the Corded Ware and Yamnaya cultures and peoples.
In a clip on Youtube he reiterated his theory that the so called Iranian-related ancestry in the Yamnaya people actually came from what is now Iran, and, more precisely, that it was carried by hunter-gatherers who travelled relatively rapidly from the South Caspian region into the Volga Delta in what is now Russia.
It's still a complete mystery to me as to why a group of hunter-gatherers from the South Caspian would undertake such a migration, instead of, say, expanding their range gradually over thousands of years, first into the Caucasus and eventually into Eastern Europe.
But there's a more serious problem with Anthony's theory: it contradicts the currently available ancient DNA. That's because the so called Iranian-related ancestry in the Yamnaya people is most closely related to the Kotias and Satsurblia hunter-gatherers from what is now Georgia, and these hunter-gatherers form a separate clade from the earliest samples from what is now Iran. For instance, see here and here.
Also, in a podcast on Razib's blog, Anthony doubled down on his theory that Y-chromosome haplogroup R1a was closely associated with Yamnaya plebs who were excluded from Kurgan burials, and, as a result, their remains haven't yet been sampled.
At least this theory isn't yet contradicted by ancient DNA, but it's more complicated and less parsimonious than my theory, which posits that R1a, or rather R1a-M417, was simply a very rare lineage in the Yamnaya population, and that it only became a common and widespread marker thanks to the Corded Ware expansion (see here).
Intriguingly, my understanding is that there are several unpublished R1a samples from the Caspian and Volga steppes at Harvard's David Reich Lab that have been classified by its scientists as Yamnaya outliers. Of course, Anthony is collaborating on at least one major paper with this lab (see here).
Ergo, I strongly suspect that Anthony's theory is in part based on these Yamnaya outliers. However, I also believe that these samples are wrongly dated and probably represent Scythians and/or Sarmatians. I'll be able to look into that if they're ever published.
Speaking of the David Reich Lab, its leading scientists, David Reich and Nick Patterson, have also made appearances online recently, on Youtube and Razib's blog, respectively, to reveal that the Corded Ware and Yamnaya peoples aren't just very similar genetically, but in fact close cousins.
This is a very interesting finding. Apparently it's based on a relatively high level of Identity-by-Descent (IBD) segment sharing between Corded Ware and Yamnaya samples, but that's all I know. I'm guessing that the relevant paper is coming soon (that is, within the next five years).
However, the long-standing question that the readers of this blog want to see answered is not whether the Corded Ware and Yamnaya peoples are close cousins, but whether Yamnaya migrants founded the Corded Ware culture. The obvious way to prove that they did is to find at least one ancient population unambiguously classified as part of the Yamnaya horizon that is rich in the typically Corded Ware Y-haplogroups R1a-M417 and R1b-L151.
See also...
On the origin of the Corded Ware people
The PIE homeland controversy: January 2019 status report
The PIE homeland controversy: August 2019 status report
Labels:
ancient DNA,
Corded Ware,
CWC,
David Anthony,
David Reich,
David Reich Lab,
Eastern Europe,
Indo-European,
Iran,
Nick Patterson,
Proto-Indo-European,
R1a-M417,
R1b-L151,
R1b-L51,
Yamna,
Yamnaya
Monday, April 26, 2021
Uralians of the Sargat horizon
Many years ago, well before the start of the ancient DNA revolution, someone made the very clever inference that the N-Tat Y-chromosome marker was closely associated with the expansion of Uralic languages.
Since then, N-Tat has been renamed several times over, to the point that I no longer know what it's called, but the aforementioned inference has turned into a very solid consensus backed up by a wide range of studies focusing on modern and ancient DNA.
Nowadays, Y-haplogroup N-L1026, a subclade of N-Tat, is seen as the main genetic signal of the Uralic expansions, along, of course, with Nganasan-related genome-wide genetic ancestry.
A recent paper at Science Advances by Gnecchi-Ruscone et al. featured the first ever genome-wide samples from the Sargat horizon, which is an Iron Age archeological formation in western Siberia normally associated with the Ugric branch of the Uralic language family. Surprisingly, and disappointingly, the authors failed to investigate this widely accepted connection.
If we go by the Y-haplogroup classifications in the paper, which may or may not be the smart thing to do, at least two of the Sargat horizon males belong to N-L1026, and one also to the more derived N-Z1936 subclade, which has been found in the remains of Hungarian Conquerers from Medieval Hungary. Of course, Hungarian is an Ugric language generally thought to have been introduced into the Carpathian Basin by the Hungarian Conquerers who originally came from western Siberia.
That's probably enough to corroborate the association between the Sargat horizon and the spread of Ugric/Uralic languages, but let's also take a quick look at the autosomal DNA of these Sargat individuals. Firstly, here's a Principal Component Analysis (PCA), based on Global25 data and produced with the Vahaduo G25 Views online tool. The results are self-explanatory.
Interestingly, I can't get a decent statistical fit when I try to reproduce the four-way qpWave/qpAdm model done by Gnecchi-Ruscone et al., probably mostly because my right pops or outgroups are different. This suggests to me that there's something important missing in their model.
Sargat_IA MNG_Khovsgol_LBA 0.203±0.045 RUS_Ekven_IA 0.183±0.044 RUS_Sintashta_MLBA 0.545±0.014 TKM_Gonur1_BA 0.068±0.013 chisq 16.805 tail prob 0.0186971 Full outputSo how about if I replace RUS_Ekven_IA with kra001, the oldest Nganasan-like individual in the ancient DNA record (see here), and MNG_Khovsgol_LBA with KAZ_Mereke_MBA, to add a more local stream of ancestry?
Sargat_IA KAZ_Mereke_MBA 0.135±0.017 kra001 0.301±0.007 RUS_Sintashta_MLBA 0.499±0.023 TKM_Gonur1_BA 0.066±0.015 chisq 8.872 tail prob 0.262001 Full outputThat's a better statistical fit and also, I'd say, a more realistic model, at least in terms of distal ancestry proportions. Note that Nganasan-related ancestry makes up 30% of the genome-wide genetic structure of the Sargat samples, which again corroborates the view that Uralic languages were spoken within the Sargat horizon. Update 28/04/21: This is the best qpAdm model that I could find for Sargat_IA, at least in terms of the chisq and tail prob. It shows that the Sargat population was in large part very similar to that of KAZ_Pazyryk_IA.
Sargat_IA KAZ_Mereke_MBA 0.032±0.016 KAZ_Pazyryk_IA 0.698±0.016 RUS_Sintashta_MLBA 0.236±0.021 TKM_Gonur1_BA 0.034±0.014 chisq 2.023 tail prob 0.958561 Full outputIt's missing kra001, because KAZ_Pazyryk_IA packs enough kra001-related ancestry for the job.
KAZ_Pazyryk_IA KAZ_Mereke_MBA 0.144±0.018 kra001 0.429±0.008 RUS_Sintashta_MLBA 0.378±0.026 TKM_Gonur1_BA 0.049±0.018 chisq 8.899 tail prob 0.259983 Full outputThe fact that KAZ_Pazyryk_IA can be modeled with significant kra001-related ancestry isn't surprising, considering that its territory was located in Siberia. However, my model doesn't necessarily prove that the Sargat population was largely or even partly of Pazyryk origin. Indeed, N-L1026 hasn't yet appeared in any Pazyryk remains. See also... The Uralic cline with kra001 - no projection this time First taste of Early Medieval DNA from the Ural region Hungarian Conquerors were rich in Y-haplogroup N More on the association between Uralic expansions and Y-haplogroup N It was always going to be this way On the association between Uralic expansions and Y-haplogroup N
Labels:
ancient DNA,
Corded Ware,
Indo-European,
Indo-Iranian,
kra001,
N-tat,
N1c,
N1c1a1a,
Nganasan,
Proto-Uralic,
R1a,
Saka,
Sargat,
Siberia,
Sintashta,
Ural,
Uralian,
Uralic
Friday, November 13, 2020
Fatyanovo as part of the wider Corded Ware family (Nordqvist and Heyd 2020)
There's a new archeological paper about the Fatyanovo culture at the Proceedings of the Prehistoric Society [LINK]. It includes this quote on page 18:
In the traditional narrative, the Fatyanovo people – like the CWC populations in general – are regarded as Indo-European, representing the pre-Balto-Slavic (-Germanic) stage (Carpelan & Parpola 2001, 88; Anthony 2007, 380; also Gimbutas 1956, 163; Tretyakov 1966, 109) in the spread of Indo-European languages.That's correct, but considering the latest ancient DNA research on the Fatyanovo people, the traditional narrative is probably wrong. Fatyanovo males were rich in Y-haplogroup R1a-Z93, which is found at very low frequencies in Balto-Slavic populations (see here). It's actually much more common nowadays in Central and South Asia, where it often reaches frequencies of over 50% in Indo-Iranian speaking groups. Balts and Slavs are rich in R1a-Z282, which is a sister clade of R1a-Z93 that has been found in Corded Ware and Corded Ware-related samples from west of Fatyanovo sites. That is, in present-day Poland and the Baltic states. Therefore, the origins of the Balto-Slavs should be sought somewhere west of the Fatyanovo culture, probably in the Corded Ware derived populations from what is now the border zone between Poland, Belarus and Ukraine. Indeed, in my view the Fatyanovo people are more likely to have spoken Proto-Indo-Iranian rather than anything ancestral to Baltic or Slavic (see here). Nordqvist and Heyd, The Forgotten Child of the Wider Corded Ware Family: Russian Fatyanovo Culture in Context, Proceedings of the Prehistoric Society, online 12 November 2020, DOI: https://doi.org/10.1017/ppr.2020.9 See also... The oldest R1a to date
Labels:
Balto-Slavic,
Balts,
Corded Ware,
Corded Ware Culture,
East Baltic,
Eastern Europe,
Fatyanovo-Balanovo culture,
Indo-European,
Poland,
Proto-Indo-European,
R1a-M417,
R1a-Z282,
R1a-Z93,
Slavs
Friday, April 17, 2020
Corded Ware cultural and genetic complexity (Linderholm et al. 2020)
Open access at Scientific Reports at this LINK. Although very useful and broadly accurate, I'm really not sure what to make of this paper yet, especially in regards to its more nuanced inferences. I'll need to look at the genotype data at some point. Worthy of note is that most of the Corded Ware males sampled by the authors belong to Y-haplogroup R1b-M269, rather than R1a-M417, which is the dominant Y-haplogroup in previously published Corded Ware samples. From the paper:
During the Final Eneolithic the Corded Ware Complex (CWC) emerges, chiefly identified by its specific burial rites. This complex spanned most of central Europe and exhibits demographic and cultural associations to the Yamnaya culture. To study the genetic structure and kin relations in CWC communities, we sequenced the genomes of 19 individuals located in the heartland of the CWC complex region, south-eastern Poland. Whole genome sequence and strontium isotope data allowed us to investigate genetic ancestry, admixture, kinship and mobility. The analysis showed a unique pattern, not detected in other parts of Poland; maternally the individuals are linked to earlier Neolithic lineages, whereas on the paternal side a Steppe ancestry is clearly visible. We identified three cases of kinship. Of these two were between individuals buried in double graves. Interestingly, we identified kinship between a local and a non-local individual thus discovering a novel, previously unknown burial custom. ... The PCA revealed that despite geographical proximity there is a distinct genetic separation between CWC and BBC individuals from southern Poland. The genetic variation of CWC individuals from southern Poland overlaps with the majority of previously published CWC individuals from Germany while the eight published CWC individuals from the Polish lowland [10,11] more closely resemble BBC individuals (Fig. S21). This fact is not unexpected if we consider the CWC communities in Polish lowlands as representatives of north-western parts of the CWC world called as the Single-Grave culture (see supplementary information). The genetic variation of BBC individuals from south-eastern Poland overlaps with the broad variation of BBC individuals from Central Europe (Bohemia, Moravia, Germany, south-western Poland and Hungary) (Fig. S22) which corresponds well with archaeological data.Linderholm, A., Kılınç, G.M., Szczepanek, A. et al. Corded Ware cultural complexity uncovered using genomic and isotopic analysis from south-eastern Poland. Sci Rep 10, 6885 (2020). https://doi.org/10.1038/s41598-020-63138-w See also... The Battle Axe people came from the steppe Is Yamnaya overrated? Single Grave > Bell Beakers
Sunday, February 17, 2019
On Maykop ancestry in Yamnaya
What Maykop ancestry in Yamnaya? There is none, or at least not enough worth discussing, except in one highly unusual female outlier from a burial in what is now eastern Ukraine. But apparently this is still up for debate? Well it shouldn't be.
To anyone with even a passing interest in the Yamnaya culture, it should be rather obvious that it formed during the tail end of the Eneolithic on the Pontic-Caspian steppe, as basically a direct offshoot of the earlier Repin culture, but perhaps also with significant influences from the earlier still Khvalynsk and Sredny Stog cultures. So why should its population history be much different from this?
It isn't, and this is fairly easy to demonstrate now despite the still rather poor sampling of Eneolithic remains from the Pontic-Caspian steppe.
Below is a series of qpAdm analyses in which I modeled several Yamnaya groups, as well as the closely related Afanasievo and Poltavka populations, exclusively and successfully as two- and three-way mixtures of a few Eneolithic singletons from various parts of the Pontic-Caspian steppe (obviously, I'd love to use homogeneous population sets instead, but, as per my point above, that's not possible yet). The models are sorted by their statistical fits, best to worst. Also note the large number and wide range of right pops or outgroups. I wanted to make sure that I wasn't missing anything.
Yamnaya_Samara Dereivka_I_I4110 0.324±0.035 Progress_Eneolithic_PG2004 0.676±0.035 chisq 6.797 tail prob 0.976979 Full output Afanasievo Progress_Eneolithic_PG2004 0.638±0.038 Sredny_Stog_II_I6561 0.362±0.038 chisq 10.855 tail prob 0.818366 Full output Yamnaya_Ukraine Progress_Eneolithic_PG2001 0.655±0.073 Sredny_Stog_II_I6561 0.345±0.073 chisq 12.676 tail prob 0.696277 Full output Poltavka Dereivka_I_I4110 0.324±0.038 Progress_Eneolithic_PG2004 0.676±0.038 chisq 12.895 tail prob 0.680437 Full output Yamnaya_Caucasus Khvalynsk_Eneolithic_I0122 0.086±0.054 Sredny_Stog_II_I6561 0.221±0.070 Vonyuchka_Eneolithic_VJ1001 0.693±0.101 chisq 13.113 tail prob 0.593562 Full outputSo, you might ask, is there any way to add Maykop to these models? Nope, it's pointless, because it doesn't improve the stats (for instance, see here, here and here). In other words, the situation is this: I already have awesome models, and I can't readily fit Maykop into my framework, so why do it? But if anyone out there wants to try, then by all means, and feel free to share the results with us in the comments. Of course, the fact that most of these Yamnaya and Yamnaya-related populations are best modeled with somewhat different Eneolithic steppe singletons doesn't mean that they have radically different origins. In fact, they're all very closely related and they're basically like one Bronze Age steppe family. They just harbor somewhat different ratios of the same ancient ancestral components. For the sake of being thorough, as per scientific literature, I pooled all of the above Afanasievo, Poltavka and Yamnaya samples into a Steppe_EMBA set and analyzed it with several genetically and geographically matching pairs of the Eneolithic singletons. This was one of the best fitting models, which I think is interesting, because the region roughly between the burial sites of these pairs of Eneolithic individuals was the home of the Repin culture.
Steppe_EMBA North_Pontic_Eneolithic_I4110-I656 0.313±0.027 Progress_Eneolithic_PG2001-PG2004 0.687±0.027 chisq 15.378 tail prob 0.497157 Full outputAgain, adding Maykop to this model makes no sense (see here, here and here). Clearly, I'd have to come up with a very different framework to successfully model Steppe_EMBA with a Maykop population. However, it's unlikely that such a model would make much sense in the context of various other types of genetic analyses and archeological data. See also... Yamnaya: home-grown Big deal of 2018: Yamnaya not related to Maykop Yamnaya isn't from Iran just like R1a isn't from India
Labels:
admixture,
Afanasievo,
ancestry,
ancient DNA,
Caucasus,
Corded Ware,
Dereivka,
Khvalynsk,
Maykop,
Poltavka,
Pontic-Caspian steppe,
R1a-M417,
R1b-M269,
Repin,
Sredny Stog,
Steppe Maykop,
Yamnaya
Tuesday, November 20, 2018
Yamnaya: home-grown
I have some interesting news. It looks like Khvalynsk_Eneolithic I0434 can be used as essentially a perfect proxy for the Eneolithic steppe trio from Wang et al. 2018 when modeling the ancestry of the Yamnaya people of what is now the Samara region of Russia. Consider the qpAdm mixture models below, sorted by taildiff.
One of the best fitting models that also fairly closely matches archeological data, which suggest that Yamnaya was an amalgamation of the Khvalynsk, Repin and Sredny Stog cultures, is in bold. The worst fitting, and basically failed, models are listed below the dotted line. Note that almost all of these models feature reference populations from West and Central Asia.
Khvalynsk_I0434 + Iberia_ChL 0.681534184 > full output Khvalynsk_I0434 + Globular_Amphora 0.525961242 > full output Khvalynsk_I0434 + Iberia_Central_CA 0.515960444 > full output Khvalynsk_I0434 + Sredny_Stog_I6561 0.485311962 > full output Khvalynsk_I0434 + Varna 0.430411416 > full output Khvalynsk_I0434 + Blatterhole_MN 0.328782809 > full output Khvalynsk_I0434 + Baden_LCA 0.234307235 > full output Khvalynsk_I0434 + Protoboleraz_LCA 0.231310724 > full output Khvalynsk_I0434 + ALPc_MN 0.200002422 > full output Khvalynsk_I0434 + Trypillia 0.193900977 > full output Khvalynsk_I0434 + Balaton_Lasinja_CA 0.187031564 > full output Khvalynsk_I0434 + Tiszapolgar_ECA 0.153940224 > full output Khvalynsk_I0434 + Tisza_LN 0.145465993 > full output Khvalynsk_I0434 + Balkans_ChL 0.111720163 > full output ... Khvalynsk_I0434 + Armenia_EBA 0.0108890099 > full output Khvalynsk_I0434 + Armenia_ChL 0.00882375703 > full output Khvalynsk_I0434 + Levant_BA_North 0.0078751978 > full output Khvalynsk_I0434 + Minoan_Lasithi 0.0675240088 > full output Khvalynsk_I0434 + Peloponnese_N 0.046998906 > full output Khvalynsk_I0434 + Hajji_Firuz_ChL 0.00269860335 > full output Khvalynsk_I0434 + Shahr_I_Sokhta_BA1 0.00261908387 > full output Khvalynsk_I0434 + Sarazm_Eneolithic 0.00120345503 > full output Khvalynsk_I0434 + Seh_Gabi_ChL 0.00111898703 > full output Khvalynsk_I0434 + Geoksiur_Eneolithic 0.000178295163 > full output Khvalynsk_I0434 + Tepe_Hissar_ChL 0.000163698274 > full output Khvalynsk_I0434 + Bustan_BA 0.000151088148 > full outputWhy is this potentially important? Because unless Khvalynsk_Eneolithic I0434 was a recent migrant from the North Caucasus piedmont steppe, which is where the remains of the Eneolithic steppe trio were excavated, then Yamnaya's ethnogenesis might not have anything at all to do with Asia or even the Caucasus region. At least not within any reasonable time frame anyway. Here's a map showing the geographic locations of all of the populations relevant to the highlighted mixture model above. I won't be fussed if it turns out that the majority of the ancestry of the Yamnaya, Corded Ware and other closely related ancient peoples was sourced from the Eneolithic populations of the North Caucasus piedmont steppe. But I think it's useful to make the point that there are still very few ancient samples available from the steppes between the Black and Caspian seas, so we don't yet have much of a clue how the groups living throughout this region during the Eneolithic and earlier fit into the grand scheme of things. Update 24/12/2018: I decided to repeat the analysis, but this time with Caucasus Hunter-Gatherers (CHG) as one of the outgroups (or right pops). The reason I initially didn't include CHG in the outgroups was because I didn't want to discriminate, perhaps unfairly, against West and Central Asians with high levels of CHG-related ancestry, and in favor of Europeans with no or minimal CHG-related input. But in my opinion, the new results clearly make more sense, with Sredny Stog and Varna at the top of the list.
Khvalynsk_I0434 + Sredny_Stog_I6561 0.410719649 > full output Khvalynsk_I0434 + Varna 0.394089365 > full output Khvalynsk_I0434 + Iberia_ChL 0.16554258 > full output Khvalynsk_I0434 + Globular_Amphora 0.128348823 > full output Khvalynsk_I0434 + Iberia_Central_CA 0.126100242 > full output Khvalynsk_I0434 + Trypillia 0.135306664 > full output Khvalynsk_I0434 + Baden_LCA 0.0853031796 > full output Khvalynsk_I0434 + Protoboleraz_LCA 0.0766892008 > full output Khvalynsk_I0434 + Tisza_LN 0.0661622403 > full output Khvalynsk_I0434 + Tiszapolgar_ECA 0.0626469042 > full output Khvalynsk_I0434 + Balaton_Lasinja_CA 0.0536293042 > full output Khvalynsk_I0434 + ALPc_MN 0.0505788809 > full output ... Khvalynsk_I0434 + Minoan_Lasithi 0.0439451605 > full output Khvalynsk_I0434 + Balkans_ChL 0.0436885241 > full output Khvalynsk_I0434 + Blatterhole_MN 0.0329758292 > full output Khvalynsk_I0434 + Peloponnese_N 0.0181930605 > full output Khvalynsk_I0434 + Armenia_EBA 0.014715999 > full output Khvalynsk_I0434 + Armenia_ChL 0.0060437014 > full output Khvalynsk_I0434 + Levant_BA_North 0.00514574731 > full output Khvalynsk_I0434 + Shahr_I_Sokhta_BA1 0.00350059625 > full output Khvalynsk_I0434 + Hajji_Firuz_ChL 0.00228771991 > full output Khvalynsk_I0434 + Seh_Gabi_ChL 0.00117061206 > full output Khvalynsk_I0434 + Sarazm_Eneolithic 0.001118931 > full output Khvalynsk_I0434 + Bustan_BA 0.00021203609 > full output Khvalynsk_I0434 + Tepe_Hissar_ChL 0.000200643323 > full output Khvalynsk_I0434 + Geoksiur_Eneolithic 0.000175941977 > full outputUpdate 17/02/2019: I basically managed to confirm my analysis with samples from the Wang et al. Caucasus paper. See here. See also... Big deal of 2018: Yamnaya not related to Maykop "The Homeland: In the footprints of the early Indo-Europeans" time map Late PIE ground zero now obvious; location of PIE homeland still uncertain, but...
Labels:
admixture,
Afanasievo,
ancestry,
ancient DNA,
Caucasus,
Corded Ware,
Dereivka,
Khvalynsk,
Maykop,
Poltavka,
Pontic-Caspian steppe,
R1a-M417,
R1b-M269,
Repin,
Sredny Stog,
Steppe Maykop,
Yamnaya
Friday, April 13, 2018
On the doorstep of India
One of the most remarkable discoveries in the recent Narasimhan et al. 2018 preprint has to be the presence of what are essentially Eastern European migrant populations within the Inner Asian Mountain Corridor (IAMC) during the Middle to Late Bronze Age (MLBA). Remarkable for so many reasons, but seemingly under-appreciated by a lot of people, judging by the online discussions that I've seen about the preprint, and even, I'd say, the authors themselves.
Narasimhan et al. labeled these groups as belonging to the "forest/steppe MLBA" complex (for instance, see the main figure from the preprint here). This is indeed what they are in terms of their genetic structure, but certainly not geography, because the IAMC is well south of the steppe. Thus, in my Principal Component Analysis (PCA) I'm going to label them as part of the "post-steppe herder expansion Turan" complex.
Strikingly, most of these people cluster with Bronze Age Eastern Europeans, and even some Bronze Age Central Europeans. They're also sitting very close to the more easterly present-day Slavic-speakers from Russia and Ukraine, and indeed closer to the bulk of the European cluster than some present-day Turkic and Uralic groups from the Volga-Ural region. Even I never predicted such an outcome. Sure, I was expecting to see ancient genomes from South Central Asia with some very heavy steppe influence, but not this. The relevant datasheet is available here.
Two of the MLBA IAMC individuals are from Kashkarchi in the Ferghana Valley, in what is now Uzbekistan, and basically on the doorstep of the Indian subcontinent. I've made special mention of them on the plot, and I've also highlighted a pair of individuals from the Bronze Age Central Asian sites of Gonur Tepe and Shahr-i Sokhta, who are, in all likelihood, unadmixed migrants from the Indus Valley (for more on that, see here).
It's surely not a coincidence that the ancient and present-day South Asians on the plot (including those from Pakistan's Swat Valley dated to the Iron Age) form an almost prefect cline between these two pairs of individuals. It's also surely not a coincidence that the MLBA IAMC groups are rich in Y-haplogroup R1a-M417, and in particular its R1a-Z93 subclade, which is today an especially frequent marker in Indo-European-speaking South Asians.
Forget about the pre-MLBA populations from the forests, steppe, or IAMC, like those represented by Dali_EBA; they're practically irrelevant to this story. How do I know? Because they have little to no impact on the above mentioned cline. And this can be easily verified with mixture models based on multiple Principal Components (PCs) and formal statistics (for instance, see here).
Clearly, many populations in South Asia, particularly those speaking Indo-European languages, derive the bulk of their steppe-related ancestry from the peoples of the MLBA IAMC, and/or their very close relatives. And if you do believe that this inference is just based on coincidences, then I'm sorry to say this, but obviously a new, much less mentally challenging, hobby or profession beckons. All the best with that.
Just to help put all of this in a geographic perspective, here's a topographical map of Eurasia. I've marked the location of the Ferghana Valley. The close relatives of Kashkarchi_BA most likely skirted their way around those winding high mountains and slipped into India via the Khyber Pass, which I've also marked on the map.
And the rest, as they say, is history, including the history described in the ancient Indo-Aryan Sanskrit texts known as the Vedas. I'm sure we'll soon be learning about these events in great detail when many more ancient samples from Pakistan and, hopefully, the first ancient samples from India, are published.
Citation...
Narasimhan et al, The Genomic Formation of South and Central Asia, Posted March 31, 2018, doi: https://doi.org/10.1101/292581
See also...
Late PIE ground zero now obvious; location of PIE homeland still uncertain, but...
Labels:
AIT,
ancient DNA,
Aryan,
Aryan Invasion Theory,
BMAC,
Corded Ware,
Eastern Europe,
India,
Indo-Aryan,
Indo-European,
Indo-Iranian,
Out-of-India,
PIE,
Pontic-Caspian steppe,
R1a,
R1a-M417,
R1a-Z93,
South Asia,
Yamnaya
Wednesday, December 31, 2008
Best of 2008: Corded Ware DNA from Germany
One of the biggest hits of the year for this blogger was the discovery of Y-DNA haplogroup R1a among three Corded Ware skeletons from a burial site in Eulau, eastern Germany. It's an important result, because it links one of Europe's most dominant Y-haplogroups to a major Late Neolithic archeological complex.
All three individuals were confirmed to be paternally related via their shared Y-STR haplotype. Nevertheless, the outcome appears far from a random coincidence. Consider that in Europe today R1a shows its highest frequencies in Poland and Western Russia, which are both located in former Corded Ware territory, and where the Eulau R1a haplotype appears to have its closest modern matches. Moreover, the Corded Ware culture is often classified as an Indo-European culture by archeologists and linguists, while at the same time R1a has been posited as a marker of the early Indo-Europeans by some geneticists. Needless to say, I'm expecting R1a to be a common, and perhaps dominant marker among Corded Ware samples when more of them make it to the lab.
The consensus haplotype of the three individuals (based on most complete profile) gave two exact matches in in an European population sample of 11,213 haplotypes in a set of 100 populations (as of July 2008, Release ‘‘23’’ from 2008–01-15 14:44:25): one individual from Poland (1/939 from Gdansk) and one from Russia (1/48 from Tambov).
...
The Y haplotype was predicted using the Web-based program Haplotype Predictor (9). The three individuals of grave 99 belong to haplotype R1a, with a probability of 100% based on the Y-STR profile of individual 3 (10). To confirm haplogroup status, we further amplified an 85-bp fragment covering the Y-SNP marker SRY10831.2 characteristic for R1a (11). Primer sequences are given in Table S6. Sequences and sequenced clones from independent extract of all three individuals show the specific G to A transition identifying R1a (Fig. S5).
The mitochondrial DNA (mtDNA) lineages of the Eulau skeletons belonged to haplogroups K1b (3), X2 (2), H, I, K1a2, and U5b. Most of these maternal markers aren't particularly common in Europe today, and the overall result appears decidedly unusual compared to the mtDNA frequencies of modern European populations, largely because of the low frequency of H.
I'm quite certain this is at least partly due to the small sample size and presence of several related individuals skewing some of the frequencies. However, it's interesting to note that this pattern of discontinuity between mtDNA gene pools from different time periods has also been reported in other studies, some with larger samples, and focusing on different regions of Europe. So it might well be a signal of significant shifts in mtDNA frequencies during European prehistory and early history, possibly as a result of major migrations leading to significant population replacements.
Interestingly, one of the ancient K1b lineages most closely matched a haplotype shared by two modern Shugnans from Tajikistan. Exactly how the Corded Ware individual is related to these two Central Asians isn't clear yet, but Shugni is an Indo-Iranian language, so some kind of early Indo-European relationship is possible.
Citation...
Wolfgang Haak et al, Ancient DNA, Strontium isotopes, and osteological analyses shed light on social and kinship organization of the Later Stone Age, PNAS, Published online before print November 17, 2008, doi:10.1073/pnas.0807592105
Subscribe to:
Posts (Atom)