This year has already been massive in all sorts of ways, including for new data and software releases. So I'm thinking it might be time to update many of the analyses that were featured at this blog a while ago.
Let's start with the classic hunter vs farmer vs herder mixture model for present-day European populations. The rules of the game are as follows:
- run the latest version of qpAdm using qpfstats output
- use transversion sites and 1240K capture data
- pick a set of diverse and chronologically sound outgroups
- for a model to be successful the p-value must reach 0.01
- tweak the left pops in models that are clearly underperforming
- follow high end scientific literature, logic and common sense
Obviously, the reason that I decided to limit my analysis to markers from transversion sites is to mitigate problems associated with modeling the ancestry of modern, high quality samples with relatively low quality ancients. One of these problems appears to be qpAdm assigning faux East Asian/Siberian admixture to present-day Europeans (for instance, see figure 4
here).
My starting reference populations and outgroups are listed below. In qpAdm terminology the former are known as the "left pops", while the latter as the "right pops". Most of these samples are freely available at the David Reich Lab website
here.
left pops:
HUN_Koros_N_HG
TUR_Barcin_N
UKR_Yamnaya
right pops:
CMR_Shum_Laka_8000BP
MAR_Taforalt
Levant_Natufian
IRN_Ganj_Dareh_N
Levant_PPNB
CZE_Vestonice16
BEL_GoyetQ116-1
Iberia_ElMiron
RUS_Karelia_HG
RUS_West_Siberia_HG
MNG_North_N
RUS_Ust_Kyakhta
As you can see, I picked a wide variety of right pops. But I chose most of them specifically to be able to differentiate the three streams of ancestry - from ancient hunters, farmers and herders - that are the focus of my analysis. I also intentionally avoided using samples in the right pops that may have experienced gene flow, including cryptic gene flow, from the populations in the left pops.
I somewhat speculatively earmarked HUN_Koros_N_HG, from the Early Neolithic Carpathian Basin, and UKR_Yamnaya, from the Early Bronze Age North Pontic steppe in what is now Ukraine, to represent the hunter-gatherer and pastoralist streams of ancestry, respectively.
That's because I expected HUN_Koros_N_HG to be the best proxy for the hunter-gatherer ancestry that was initially absorbed by the early farmers who fanned out from the Aegean region across much of the European continent, and of course it made sense to choose a steppe pastoralist population that was located close to Central Europe where such groups first made the biggest impact outside of the steppe.
Interestingly, HUN_Koros_N_HG and UKR_Yamnaya did prove to be among most effective choices for the types of ancestries that they represented. For instance, UKR_Yamnaya generally produced much stronger statistical fits than a very similar set of Yamnaya samples from the Caspian steppe (more precisely, from the Samara region in Russia). However, this might well be an artifact, due to very specific characteristics of these few ancient individuals. Larger sample sets would be welcome, especially from Yamnaya sites in Ukraine.
Below, dear audience, is a spreadsheet featuring the preliminary results. Click on the image to view and/or download the spreadsheet. The general rule is that the higher the tail prob, or p-value, the more likely it is that the ancestry proportions are close to the truth (a tail prob of well below 0.05 is usually a strong indication that something isn't right). For a detailed look at each of the qpAdm runs, feel free to consult the zip file
here.
Note, however, that many of the European groups in my burgeoning genotype dataset are yet to make an appearance in the spreadsheet. That's because their models with the standard left pops showed p-values well under 0.01, which essentially meant that they failed, and I'm still trying to make them work.
But round one has certainly revealed some fascinating stuff. For instance, except for Hungarians and Estonians, none of the Uralic-speaking groups can be modeled successfully in the standard three-way model.
However, I managed to significantly improve the statistical fits in their models by adding a Siberian population, RUS_Baikal_BA, to the left pops. This is unlikely to be a coincidence, because the Proto-Uralic homeland was almost certainly located in or very near Siberia.
Iain Mathieson please take note.
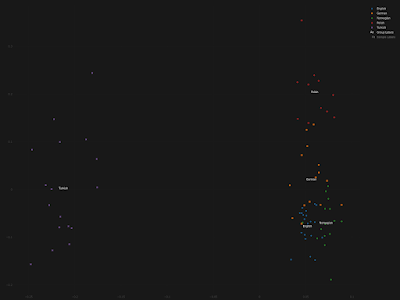
Saami
HUN_Koros_N_HG 0.134±0.043
RUS_Baikal_BA 0.270±0.015
TUR_Barcin_N 0.081±0.026
UKR_Yamnaya 0.515±0.058
chisq 19.865
tail prob 0.0108571
See also...