Ancient DNA has revealed that large scale migrations and population replacements have often accompanied major cultural changes in prehistoric Europe. But, for now, my opinion is that the formation of the archeologically ostentatious Nordic Bronze Age wasn't associated with any significant foreign gene flow into Scandinavia. I've tested this as best as I could with the few relevant ancient samples that are currently available.
For instance, below are among the most successful qpAdm mixture models that I was able find for various ancient Scandinavian groups dating back to the local Middle Neolithic (MN) period. The Nordic Bronze Age population is represented by three individuals labeled Nordic_BA. Unfortunately, the guy pictured above, from the famous Borum Eshøj barrow burial in what is now Denmark, didn't make the cut. For more details about my sampling and labeling strategies refer to the text file
here.
Nordic_MN_B
CWC_CZE 0.822±0.059
POL_Globular_Amphora 0.178±0.059
chisq 14.478
tail prob 0.341086
Full output
SWE_Battle_Axe
CWC_Baltic_early 0.662±0.028
POL_Globular_Amphora 0.338±0.028
chisq 11.234
tail prob 0.591189
Full output
Nordic_LN
Nordic_MN_B 0.928±0.069
SWE_TRB 0.072±0.069
chisq 12.139
tail prob 0.516307
Full output
Nordic_BA
Nordic_LN 0.851±0.061
SWE_TRB 0.149±0.061
chisq 10.897
tail prob 0.619475
Full output
It's impossible to successfully model the ancestries of Nordic_MN_B and SWE_Battle_Axe simply with the populations that were living in Scandinavia before them. Therefore, it's likely that they were migrants or the recent descendants of migrants to Scandinavia. But there's nothing surprising about that, because they're archeologically associated with the Corded Ware culture (CWC), which has always been seen as intrusive to Scandinavia from the south and east.
Conversely, it's easy to produce statistically sound mixture models for both Nordic_LN and Nordic_BA exclusively with earlier Scandinavian populations. Indeed, based on the outgroups or right pops that I'm using, Nordic_LN is almost indistinguishable from Nordic_MN_B, and the same can be said of Nordic_BA in regards to Nordic_LN.
Of course, if I mixed and matched reference populations from across prehistoric Europe, I could probably come up with some spectacular statistical fits even without the need for any Scandinavians. Essentially that's because Nordic_LN and Nordic_BA are closely related to many earlier and contemporaneous peoples living all the way from the Atlantic facade to the Ural Mountains. My point, however, is that this isn't crucial, despite the dearth of ancient samples from Scandinavia.
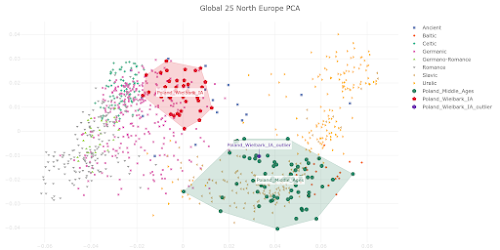
This is how things look in a Principal Component Analysis (PCA) of Northern European genetic variation based on my
Global25 data. Strikingly, Nordic_MN_B, SWE_Battle_Axe, Nordic_LN and Nordic_BA more or less recapitulate the cluster made up of present-day Swedish samples. The relevant datasheet is available
here.
Granted, two of the Nordic_BA samples sit just south of the Swedes, no doubt due to their slightly higher ratios of Neolithic farmer (SWE_TRB-related) ancestry, but this is also an area of the plot that many present-day Danes call home (not shown, because I don't have any suitable academic Danish samples to run).
I'll eat my hat if it turns out that Scandinavia experienced a major population shift (say, more than a collateral ~10%) during the LN and/or BA periods. And I'll post a clip of it online too.
Update 27/08/2019: Four of the samples from the recent
Frei et al. paper on human mobility in prehistoric southern Scandinavia are in my
Global25 datasheets. So I thought it might be interesting to check whether their strontium isotope ratios correlated with their genomic profiles.
In the Principal Component Analysis (PCA) below, RISE61 is a subtle outlier along the horizontal axis compared to the other three Nordic ancients, as well as a Danish individual representative of the present-day Danish gene pool. Also note that RISE61 shows the most unusual strontium isotope ratio (0.712588). The PCA was run with an online tool freely available
here.
To help drive the point home, here's a figure from Frei et al., edited by me to show the positions of RISE47, RISE61 and RISE71. If RISE276 was also in this graph, he'd be sitting well under the "local" baseline, in roughly the same spot along the vertical axis as RISE47.
Interestingly, RISE61 belongs to Y-chromosome haplogroup R1a-M417, while RISE47 and RISE276, who appear to have been locals, both belong to R1b-M269. My guess is that RISE61 was a recent migrant from a more northerly part of Scandinavia dominated by the Battle-Axe culture (BAC). The BAC population was probably rich in R1a-M417 because it moved into Scandinavia from the Pontic-Caspian steppe via the East Baltic. This is what Frei et al. say about RISE61 and his burial site:
The double passage grave of Kyndeløse (Fig 1, S1 File) located on the island of Zealand yielded 70 individuals as well as a large number of grave goods, including flint artefacts, ceramics, and tooth and amber beads. We conducted strontium isotope analyses of seven individuals from Kyndeløse encompassing a period of c. 1000 years, indicating the prolonged use of this passage grave. The oldest of the seven individuals is a female (RISE 65) from whom we measured a “local” strontium isotope signature ( 87 Sr/ 86 Sr = 0.7099). Similar values were measured in five other individuals, including adult males and females. Only a single individual from Kyndeløse, an adult male (RISE 61) yielded a somewhat different strontium isotope signature of 87 Sr/ 86 Sr = 0.7126 which seems to indicate a non-local provenance. The skull of this male individual revealed healed porosities in the eye orbits, cribra orbitalia, a condition which is possibly linked to a vitamin deficiency during childhood, such as iron deficiency.
By the way, RISE47 was buried in a flat grave, which suggests that he was a commoner. RISE276 was found in a peat bog in Trundholm, where the famous Trundholm sun chariot was discovered (see
here). He may have been a human sacrifice.
Citation...
Frei KM, Bergerbrant S, Sjögren K-G, Jørkov ML, Lynnerup N, Harvig L, et al. (2019)
Mapping human mobility during the third and second millennia BC in present-day Denmark. PLoS ONE 14(8): e0219850. https://doi.org/10.1371/journal.pone.0219850
See also...
They came, they saw, and they mixed
Children of the Divine Twins
The mystery of the Sintashta people