When used properly, Principal Component Analysis (PCA) is an extraordinarily powerful tool and one of the best ways to study fine-scale genetic substructures within Europe.
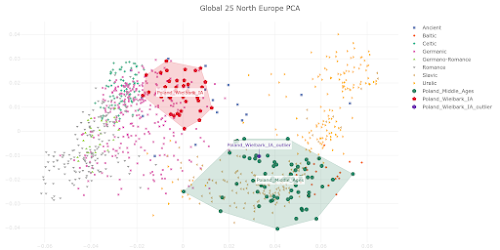
The PCA plot below is based on Global25 data and focuses on the genetic relationship between Wielbark Goths and Medieval Poles, including from the Viking Age, in the context of present-day European genetic variation.
I'd say that it's a wonderfully self-explanatory plot, but here are some key observations:
- the Wielbark Goths (Poland_Wielbark_IA) and Medieval Poles (Poland_Middle_Ages) are two distinct populations
- moreover, the Wielbark Goths form a relatively compact Scandinavian-related cluster and must surely represent a homogenous population overwhelmingly of Scandinavian origin
- on the other hand, the Medieval Poles form a more extensive and heterogeneous cluster that overlaps with present-day groups all the way from Central Europe to the East Baltic, and that's because they are likely to be in large part of mixed origin
- I know for a fact that at least some of these early Poles harbor recent admixture, because their burials are similar to those of Vikings and their haplotypes have been shown to be partly of Scandinavian origin (see here)
- one of the Wielbark females is an obvious genetic outlier (Poland_Wielbark_IA_outlier), and basically looks like a first generation mixture between a Goth and a Balt.
Please note that the PCA is only based on relatively high quality genomes, so as not to confuse the picture with spurious results and noise. Also, all outliers with potentially significant ancestry from outside of Central, Eastern and Northern Europe were removed from the analysis. The relevant datasheet is available
here.
However, sanity checks are always important when studying complex topics like fine-scale genetic ancestry. To that end I've prepared a graph based on f3-statistics of the form f3(X,Cameroon_SMA,Estonia_BA)/(X,Cameroon_SMA,Ireland_Megalithic), that reproduces the key features of my PCA. The relevant datasheet is available
here.
Polish groups from the Middle Ages are marked with the MA suffix, while the Iron Age Wielbark Goths are marked with the IA suffix.
If you're wondering why I plotted the f3-statistics that I did, take a look at this (all groups largely of Scandinavian origin are emboldened):
f3(X,Estonia_BA,Cameroon_SMA)
Poland_Legowo_MA 0.226406
Poland_Ostrow_Lednicki_MA 0.225996
Poland_Plonsk_MA 0.225017
Poland_Trzciniec_Culture 0.224215
Poland_Lad_MA 0.224142
Poland_Viking 0.223838
Poland_Niemcza_MA 0.223659
Poland_Weklice_IA 0.223549
Poland_Kowalewko_IA 0.222584
Poland_Pruszcz_Gdanski_IA 0.222324
Sweden_Viking 0.222091
Russia_Viking 0.222042
Poland_Maslomecz_IA 0.221914
Norway_Viking 0.221825
Denmark_EarlyViking 0.221257
Denmark_Viking 0.221174
England_Viking 0.220979
f3(X,Ireland_Megalithic,Cameroon_SMA)
Poland_Maslomecz_IA 0.219816
Poland_Weklice_IA 0.219501
Denmark_Viking 0.2192
Poland_Kowalewko_IA 0.219176
Poland_Ostrow_Lednicki_MA 0.218916
Norway_Viking 0.218854
Poland_Pruszcz_Gdanski_IA 0.218684
Sweden_Viking 0.218626
Denmark_EarlyViking 0.218529
England_Viking 0.218308
Russia_Viking 0.217999
Poland_Viking 0.217914
Poland_Plonsk_MA 0.217756
Poland_Lad_MA 0.217719
Poland_Legowo_MA 0.21765
Poland_Niemcza_MA 0.217001
Poland_Trzciniec_Culture 0.216551
Interestingly, the Middle Bronze Age samples associated with the Trzciniec Culture (Poland_Trzciniec_Culture) show a closer genetic relationship to Medieval Poles than to Wielbark Goths or Northwestern Europeans. This is indeed the case both in terms of genome-wide and uniparental markers, including some very specific lineages under Y-chromosome haplogroup R1a.
But that's a much more complex issue that I'll leave for another time. So please stay tuned.
See also...
Slavs have little, if any, Scytho-Sarmatian ancestry