- the Yamnaya individuals sit on genetic clines made up of hunter-gatherers native to the Caucasus and various parts of Eastern Europe, including a trio from the southernmost part of the Pontic-Caspian steppe (labeled Steppe_Eneolithic), with whom they form a distinct cluster - the samples from the Caucasus and the Iranian Plateau form very different clusters, so there's no support here for the ancient Caucasus/Iranian grouping that is often haphazardly invoked in scientific literature - there's no indication that the Yamnaya and/or Steppe_Eneolithic groups experienced recent gene flow, or, for that matter, any gene flow whatsoever, from what is now Iran.Of course, analyses based on formal statistics suggest that the Yamnaya population harbors minor western ancestry that is missing in Steppe_Eneolithic. In fact, I was first to argue this point (see here). So let's add a couple of ancient farmers from Western Europe to my PCA to see how they affect the graphs. The relevant datasheet is available here. Yep, the Yamnaya pair appears to be peeling away very slightly, but deliberately, from the Steppe_Eneolithic individuals towards the part of the plot occupied by the farmers. Admittedly, I'm no Sherlock Holmes, but even with my fairly average sleuthing abilities, I'm pretty sure I know how the Yamnaya people came to be. They formed largely on the base of a population very much like Steppe_Eneolithic somewhere deep in Eastern Europe, well to the north of the Caucasus, and nowhere near the Iranian Plateau. See also... A note on Steppe Maykop
search this blog
Monday, December 30, 2019
A final note for the year
I feel like I've spent a good part of 2019 banging my head against a thicker than average brick wall.
Much of this feeling is tied to the controversy over the ethnogenesis of the Yamnaya people, and my often futile attempts to explain that their origin cannot be sought in what is now Iran, or, indeed, anywhere outside of Eastern Europe.
This post is my final attempt to lay out the facts in regards to this topic. Next year I'll have better things to do than to argue the bleeding obvious.
Below are two graphs from a Principal Component Analysis (PCA) based on relatively high quality ancient human genotype data from the Caucasus and surrounds. They include two typical Yamnaya individuals from burial sites north of the Caspian Sea. I made the graphs with the Vahaduo Custom PCA tool here. The relevant datasheet can be downloaded here.
Here's what I'm seeing:
Friday, December 20, 2019
A note on Steppe Maykop
I'm reading a new book titled Dispersals and Diversification: Linguistic and Archaeological Perspectives on the Early Stages of Indo-European (see here). One of the chapters is authored by archeologist David Anthony, in which he makes the following claims:
A previously unknown genetic population actually was identified in Wang et al. (2019), but it was a peculiar relict-seeming group related to Paleo Siberians and American Indians (Kennewick) that had survived isolated somewhere in the Caspian steppes or perhaps in the North Caucasus Mountains. The Maykop people did admix with this previously isolated Siberian/Kennewick population in graves labeled "Steppe Maykop" in Wang et al. (2019). But this just makes it clearer that a cultural choice motivated the Maykop people to exclude marriages with Yamnaya and pre-Yamnaya people specifically, even while exchanges of material goods, ideas, technologies continued. Neither the Maykop nor the North Caucasus/Siberian/Kennewick population can be the source of most of the CHG [Caucasus hunter-gatherer] ancestry in Yamnaya. In order to narrow down when and where CHG ancestry entered the steppes, we must widen our geographic frame beyond the Caucasus.Unfortunately, this is way off the mark. Especially unsound is his inference that the CHG-related ancestry in the Yamnaya population may have come from beyond the Caucasus. In fact, the chances that the Steppe Maykop people were derived from a relict Siberian/Kennewick-related group that survived into the Maykop era in the Caspian steppes or the North Caucasus are exactly zero. The real story was surely more complicated. In my opinion, it initially involved the migration during the Eneolithic or earlier of a people rich in CHG ancestry from the southernmost steppes into the Volga Delta and surrounds, and then the back-migration during the Early Bronze Age (EBA) of their descendants with around 50% admixture from Central Asian foragers. If so, these foragers were very similar to indigenous West Siberians and also relatively closely related to Native Americans. I don't know why such an exotic people migrated into the North Caucasus steppes to form the bulk of the Steppe Maykop population, but I'm certain they did, and one interesting possibility is that they were recruited by Maykop chiefs to create a buffer zone against hostile Yamnaya-related groups trying to push into the Caucasus, possibly from the lower Don region. Of course, the same ancient northward migration of the CHG-rich population that may have eventually given rise to the Steppe Maykop people might also explain the deep origins of the Yamnaya people. The key sample in all of this is VJ1001 from the Wang et al. paper. This female comes from an Eneolithic (4332-4238 calBCE) kurgan burial in the North Caucasus steppes. But despite her early date, she's genetically very similar to most Yamnaya individuals. And she's also a perfect proxy for half of the ancestry of three out of the six Steppe Maykop individuals. Here's a mixture model that I put together using the Broad MIT/Harvard software qpAdm:
RUS_Steppe_Maykop (3/6) RUS_Eneolithic_steppe_VJ1001 0.452±0.023 RUS_Tyumen_HG 0.548±0.023 chisq 7.494 tail prob 0.874914 Full outputIndeed, these Steppe Maykop samples don't harbor any Maykop ancestry. They're simply a two-way mixture between a population closely resembling VJ1001 and another one similar to hunter-gatherers from Tyumen, West Siberia. Importantly, a couple of Steppe Maykop-related populations were inadvertently discovered by Narasimhan et al. northeast of the Caspian Sea in what is now Kazakhstan. One of these groups is labeled Kumsay_EBA, after the location of its cemetery. It's roughly contemporaneous with Steppe Maykop and basically identical to the aforementioned Steppe Maykop trio.
KAZ_Kumsay_EBA RUS_Eneolithic_steppe_VJ1001 0.440±0.022 RUS_Tyumen_HG 0.560±0.022 chisq 10.573 tail prob 0.646513 Full outputI suppose it's possible that Kumsay_EBA represents the migration of Steppe Maykop people into the Kazakh steppes. But even if this is true, then there had to have been an earlier migration of a group from the Kazakh steppes or West Siberia that mixed with the VJ1001-related natives of the North Caucasus steppes to give rise to Steppe Maykop. I'm assuming that the Yamnaya-like VJ1001 and her people were the indigenous population of the North Caucasus steppes because there are no indications that they or their ancestors migrated there within any reasonable time frame from anywhere else, and certainly not from as far afield as, say, what is now Iran. The other three Steppe Maykop individuals, who are genetic outliers in varying degrees from the main Steppe Makyop cluster, show variable levels of Maykop ancestry, with an average of about 50%. But they too harbor significant VJ1001-related ancestry. So despite the fact that there was some irregular mixing between the Maykop and Steppe Maykop peoples, this is not what created the typical Steppe Maykop genetic profile.
RUS_Steppe_Maykop_o RUS_Eneolithic_steppe_VJ1001 0.234±0.074 RUS_Maykop_Novosvobodnaya 0.461±0.046 RUS_Tyumen_HG 0.305±0.033 chisq 7.378 tail prob 0.831667 Full outputAnd, of course, it should be obvious by now that the ancestry of the vast majority of Yamnaya individuals is better modeled without any input whatsoever from the Maykop or Steppe Maykop samples. In fact, early indications are that the Yamnaya people flooded into Steppe Maykop territory from the north and completely replaced its population (see here). Despite this, in Dispersals and Diversification archeologist Kristian Kristiansen makes the following claim: "steppe Maykop expanded north, leading to the formation of the Yamnaya Culture and Proto-Indo-European". Not a chance in hell Professor. See also... A final note for the year The PIE homeland controversy: August 2019 status report Some myths die hard An exceptional burial indeed, but not that of an Indo-European
Labels:
ancient DNA,
Caspian Sea,
Caucasus hunter-gatherers,
Central Asia,
Eastern Europe,
Indo-European,
Iran,
Maykop,
Pontic-Caspian steppe,
R1a-M417,
R1b-M269,
Steppe Maykop,
West Siberia,
Yamnaya
Saturday, December 14, 2019
Avalon vs Valhalla revisited
Pictured below is a new version of my Celtic vs Germanic genetic map. It's based on the same Principal Component Analysis (PCA) as the original (which can be seen here), but more focused on Northwestern Europe and produced with a different program.
To see the interactive online version, navigate to Vahaduo Custom PCA and copy paste the text from here into the empty space under the PCA DATA tab. Then press the PLOT PCA button under the PCA PLOT tab. For more guidance, refer to the screen caps here and here.
To include a wider range of populations in the key, just edit the data accordingly. For instance, to break up the ancient grouping into more specific populations, delete the Ancient: prefix in all of the relevant rows. This is what you should see:
Conversely, you can leave the ancient sample set intact and instead reorder the present-day linguistic groupings into, say, geographic groupings. To achieve this just delete all of the linguistic prefixes, such as Celtic:, Germanic:, and so on. You should end up with a datasheet like this and plot like this.
Of course, you can design your own plot by using any combination of the ancient and present-day individuals and populations that I've already run in this PCA. Their coordinates are listed here. Indeed, if you're in the possession of your own Celtic vs Germanic PCA coordinates, you can add yourself to the plot. And if you're not, see here.
It's also possible to re-process PCA data via the SOURCE tab. But I don't recommend doing this with the Celtic vs Germanic data, which are derived from a fine scale analysis and don't pack much variation. On the other hand, Global25 data are ideal for such re-processing. I made the plots below from subsets of Global25 coordinates available in a zip file here. To see how, refer to the screen caps here and here.
See also...
Modeling your ancestry has never been easier
Getting the most out of the Global25
Modeling genetic ancestry with Davidski: step by step
Labels:
ancient DNA,
Anglo-Saxon,
Avalon,
British Isles,
Celtic,
Celtic vs Germanic,
Gaelic,
Germanic,
Ireland,
Irish,
Nordic,
North Sea,
Northern Europe,
Northwestern Europe,
PCA,
Scandinavia,
Valhalla,
Viking
Monday, December 9, 2019
The BOO people: earliest Uralic speakers in the ancient DNA record?
N-L1026 is the Y-chromosome haplogroup most closely associated with the speakers of Uralic languages. Thus far, the oldest published instances of N-L1026 are in two Siberian-like samples dating to 1473±87 calBCE from the site of Bolshoy Oleni Ostrov (BOO), located within the Arctic Circle in the Kola Peninsula, northern Russia.
So does this mean that the BOO people were Uralic speakers? I'm now thinking that it probably does, even though, as the scientists who published the BOO samples a year ago pointed out, they predate most estimates of the spread of extant Uralic languages into the Kola Peninsula (see Lamnidis et al. here).
Hundreds of ancient human samples from across Eurasia have been sequenced since last year. In fact, thousands if we count unpublished data. But only a handful of them belong to N-L1026.
Indeed, as far as I know, the next oldest instance of N-L1026 from Europe after those at BOO is still in an Iron Age sample from what is now Estonia published earlier this year as 0LS10. Of course, this individual was in all likelihood an early west Uralic (Finnic) speaker (see Saag et al. here).
Moreover, consider these comments by Murashkin et al. in regards to the BOO site (referred to as KOG in their paper, available here):
Most of the bodies had been buried in wooden, boat-shaped, lidded caskets, which looked like small boats or traditional Sámi sledges (Ru. kerezhka). ... The morphological characteristics of the skull series of the KOG are not like those of any other ancient or modern series from the Kola Peninsula, including the Sámi people. Instead, the series shows closer biological affinities with ancient Altai Neolithic and modern, Ugric-speaking Siberian groups (Moiseyev & Khartanovich 2012). It has earlier been suggested that modern Ugric-speaking Siberians, together with Samoyeds and Volga Finnic populations, share some common morphological characteristics that indicate their common origin (Alekseyev 1974; Bunak 1956; Gokhman 1992). ... Based on the materials from the grave field, we can argue that there were direct or indirect contacts between the inhabitants of the Kola Peninsula and southern and western Scandinavia (Murashkin & Tarasov 2013).Thus, the BOO people may have spoken an early west Uralic language related to Sami languages. It's also possible that they are in part ancestral to the N-L1026-rich Sami people. Another intriguing thing about these mysterious ancients is that individual BOO003 belongs to the rare mitochondrial haplogroup T2d1b1. Now, this clearly is not a lineage native to Europe or indeed any part of North Eurasia. Its ultimate source is probably West or Central Asia. So how did this pioneer polar explorer end up with such an unusual and exotic mtDNA marker, and might the answer be an important clue about the origins of the BOO people? The most plausible explanation is that the ancestors of BOO003 were associated with the Seima-Turbino phenomenon, which stretched from the taiga zone to the oases of what is now western China along the Ob-Irtysh river system, and probably facilitated cultural, linguistic and genetic exchanges between the populations of North Eurasia and Central Asia. In other words, considering all of the clues, it would seem that the BOO people came from some part of the Ob-Irtysh basin, which might thus be the best place to look for the population with the oldest and phylogenetically most basal N-L1026 lineages. And if we find that, then we've probably found the proto-Uralians and their homeland. Below is a Principal Component Analysis (PCA) based on Global25 data featuring the earliest likely Uralic speakers in the ancient DNA record. It was produced with an online PCA runner freely available here. EST_IA includes the above mentioned 0LS10, while FIN_Levanluhta_IA is largely made up of Saami-related samples from western Finland. See anything interesting? Feel free to let me know about it in the comments below. See also... Big deal of 2019: ancient DNA confirms the link between Y-haplogroup N and Uralic expansions It was always going to be this way More on the association between Uralic expansions and Y-haplogroup N
Labels:
ancient DNA,
Arctic,
Bolshoy Oleni Ostrov,
BOO,
Fennoscandia,
Indo-European,
Indo-Iranian,
N-L1026,
N1c,
N1c1a1a,
Northeastern Europe,
Proto-Indo-European,
Proto-Uralic,
Seima-Turbino,
Siberia,
Uralic,
Urals
Sunday, December 1, 2019
Big deal of 2019: ancient DNA confirms the link between Y-haplogroup N and Uralic expansions
The academic consensus is that Indo-European languages first spread into the Baltic region from the Eastern European steppes along with the Corded Ware culture (CWC) and its people during the Late Neolithic, well before the expansion of Uralic speakers into Fennoscandia and surrounds, probably from somewhere around the Ural Mountains.
On the other hand, the views that the Uralic language family is native to Northern Europe and/or closely associated with the CWC are fringe theories usually espoused by people not familiar with the topic or, unfortunately it has to be said, mentally unstable trolls.
The likely close relationship between the CWC expansion and the early spread of Indo-European languages was discussed in several papers in recent years (for instance, see here). This year, we saw the first ancient DNA paper focusing on the transition from the Bronze Age to the Iron Age in the East Baltic, including the likely first arrival of Uralic speech in what is now Estonia.
Published in Current Biology courtesy of Saag et al., the paper showed that the genetic structure of present-day East Baltic populations largely formed in the Iron Age (see here). It was during this time, the authors revealed, that the region experienced a sudden influx of Y-chromosome haplogroup N, which is today common in many Uralic speaking populations and often referred to as a Proto-Uralic marker. Little wonder then that Saag et al. linked this genetic shift in the East Baltic to the westward migrations of early Uralic speakers.
The table below, based on data from the Saag et al. paper, surely doesn't leave much to the imagination about what happened.
Unfortunately, I have to say that the genome-wide analysis in the paper was less informative than it could have been. The authors focused their attention on rather broad genetic components, and, as a result, missed an interesting fine scale distinction between their Bronze Age and Iron Age samples. The spatial maps below, based on my Global25 data for most of the ancients from Saag et al., show what I mean. The hotter the color the higher the genetic similarity between them and present-day West Eurasian populations.
Note that the Bronze Age (Baltic_EST_BA) samples are most similar to the Baltic-speaking, and thus also Indo-European-speaking, Latvians and Lithuanians, rather than the Uralic-speaking Estonians, even though they're from burial sites in Estonia. On the other hand, the Iron Age (Baltic_EST_IA) samples show strong similarity to a wider range of populations, including Estonians and many other Uralic-speaking groups.
See also...
It was always going to be this way
Fresh off the sledge
More on the association between Uralic expansions and Y-haplogroup N
Labels:
ancient DNA,
Corded Ware Culture,
East Baltic,
Estonia,
Fennoscandia,
Finnic,
Finno-Ugric,
Indo-European,
N1c,
N1c1a1a,
N3a,
Northeastern Europe,
Proto-Uralic,
R1a,
R1a-M417,
Siberia,
Uralic,
Urals
Monday, November 25, 2019
Viking Age Iceland
I finally managed to get some of the Icelandic ancients from Ebenesersdóttir et al. 2018 into the Global25 datasheets (see here). Better late than never. Look for the"ISL_Viking_Age" prefix. Below is a screen cap of a Principal Component Analysis (PCA) with the new samples. It was done with an online Global25 PCA runner freely available here.
The individuals classified as unadmixed Gaels and Norse by Ebenesersdóttir et al. generally also look like it based on their Global25 coordinates.
The mixture models below, using all of the populations from the Global25 "modern pop averages scaled" datasheet, were run with an online tool freely available here. Note that the ADD DIST COL option is set to 1X. This is a useful feature for modeling the fine scale ancestry of samples that are derived from very similar populations.
See also...
They came, they saw, and they mixed
Commoner or elite?
Who were the people of the Nordic Bronze Age?
Labels:
ancient DNA,
Bjork,
Celtic,
Gaelic,
Gaels,
Germanic,
Global25,
I1,
Iceland,
Icelandic,
Ireland,
Norse,
Norsemen,
Northern Europe,
PCA,
R1a-Z284,
R1b-L21,
Scandinavia,
Viking Age,
Vikings
Sunday, November 10, 2019
Etruscans, Latins, Romans and others
I've just added coordinates for more than 100 ancient genomes from the recently published Antonio et al. ancient Rome paper to the Global25 datasheets. Look for the population and individual codes listed here. Same links as always:
Global25 datasheet ancient scaled Global25 pop averages ancient scaled Global25 datasheet ancient Global25 pop averages ancientThus far I've only managed to check a handful of the coordinates, so please let me know if you spot any issues. Below is a Principal Component Analysis (PCA) featuring the Etruscan and Italic speakers. I ran the PCA with an online tool specifically designed for Global25 coordinates freely available here. Can we say anything useful about the origins of the Etruscan and early Italic populations thanks to these new genomes? Also, to reiterate my question from the last blog post, what are the genetic differences exactly between the Etruscans, early Latins, Romans and present-day Italians? Feel free to let me know in the comments below. Update 13/11/2019: Here's another, similar PCA. This one, however, is based on genotype data, and it also highlights many more of the samples from the Antonio et al. paper. Considering these results, I'm tempted to say that the present-day Italian gene pool largely formed in the Iron Age, and that it was only augmented by population movements during later periods. The relevant datasheet is available here. Update 13/11/2019: It seems to me that the two Latini-associated outliers show significant ancestry from the Levant, which possibly means that they're in part of Phoenician origin. These qpAdm models speak for themselves:
ITA_Ardea_Latini_IA_o ITA_Proto-Villanovan 0.547±0.081 Levant_ISR_Ashkelon_IA2 0.453±0.081 chisq 7.573 tail prob 0.87027 Full output ITA_Prenestini_tribe_IA_o ITA_Proto-Villanovan 0.679±0.068 Levant_ISR_Ashkelon_IA2 0.321±0.068 chisq 7.222 tail prob 0.89033 Full outputThe Proto-Villanovan singleton is also a key part of the models. Dating to the Bronze Age/Iron Age transition, she appears to be of western Balkan origin. Moreover, her steppe ancestry is probably derived directly from the Yamnaya horizon.
ITA_Proto-Villanovan HRV_Vucedol 0.677±0.031 Yamnaya_RUS_Samara 0.323±0.031 chisq 10.397 tail prob 0.661174 Full outputThe cluster made up of four early Italic speakers can be modeled with minor Proto-Villanovan-related ancestry, but, perhaps crucially, it doesn't need to be. Indeed, judging by the qpAdm output below, it's possible that almost all of its steppe ancestry came from the Bell Beaker complex, and, thus, the Corded Ware culture complex before that.
ITA_Italic_IA Bell_Beaker_Mittelelbe-Saale 0.480±0.055 ITA_Grotta_Continenza_CA 0.411±0.042 ITA_Proto-Villanovan 0.109±0.084 chisq 10.294 tail prob 0.590205 Full outputTwo out of the three available Etruscans look very similar to the Italic speakers in the above PCA plots, and yet they show a lot more Proto-Villanovan-related ancestry in my qpAdm run. The statistical fit is also relatively poor, perhaps suggesting that something important is missing.
ITA_Etruscan Bell_Beaker_Mittelelbe-Saale 0.186±0.081 ITA_Grotta_Continenza_CA 0.283±0.064 ITA_Proto-Villanovan 0.531±0.126 chisq 17.175 tail prob 0.143143 Full outputInterestingly, the Etruscan outlier with significant North African admixture (proxied in my run by MAR_LN) doesn't need to be modeled with any Bell Beaker ancestry.
ITA_Etruscan_o ITA_Proto-Villanovan 0.675±0.057 MAR_LN 0.325±0.057 chisq 14.864 tail prob 0.315912 Full outputUpdate 17/11/2019: The spatial maps below show how three groups of ancient Romans (from the Imperial, Late Antiquity and Medieval periods) compare to present-day West Eurasian populations in terms of their Global25 coordinates. The hotter the color, the higher the similarity. More here. See also... Getting the most out of the Global25
Labels:
admixture,
ancient DNA,
Ancient Greece,
Ancient Rome,
archaeogenetics,
Etruscans,
Global25,
Italic,
Italy,
Latin,
Levant,
Mediterranean,
Middle East,
North Africa,
Pontic-Caspian steppe,
Roman Empire,
Southern Europe
Thursday, November 7, 2019
What's the difference between ancient Romans and present-day Italians?
The first paper on the genomics of ancient Romans was finally published today at Science [LINK]. It's behind a paywall, but the supplementary info is freely available here. Below is a quick summary of the results courtesy of the accompanying Ancient Rome Data Explorer.
I'm told that the genotype data from the paper will be online within a day or so at the Pritchard Lab website here. I'll have a lot more to say about ancient Romans and present-day Italians after I get my hands on it.
See also...
Etruscans, Latins, Romans and others
Labels:
admixture,
ancient DNA,
Ancient Greece,
Ancient Rome,
archaeogenetics,
Etruscans,
Italian,
Italic,
Italy,
Latin,
Levant,
Mediterranean,
Middle East,
North Africa,
Pontic-Caspian steppe,
Roman Empire,
Southern Europe
Tuesday, November 5, 2019
Modeling your ancestry has never been easier
An exceedingly simple, yet feature-packed, online tool ideal for modeling ancestry with Global25 coordinates is freely available HERE. It works offline too, after downloading the web page onto your computer. Just copy paste the coordinates of your choice under the "source" and "target" tabs, and then mess around with the buttons to see what happens. The screen caps below show me doing just that.
Another free, easy to use online tool that works with Global25 coordinates is the Principal Component Analysis (PCA) runner HERE. Below is a screen cap of me checking out one of the many PCA that it offers.
See also...
Getting the most out of the Global25
Saturday, November 2, 2019
Interesting times ahead
The map below made a big impression on me. Can't wait to see all of these ancient samples online. More details here.
See also...
Is Yamnaya overrated?
Y-haplogroup R1a and mental health
Getting the most out of the Global25
Labels:
ancient DNA,
archaeogenetics,
archeology,
Britain,
Central Asia,
Eurasia,
Eurasian steppe,
Europe,
global genetic variation,
HIV,
Iberia,
Indo-European,
Pontic-Caspian steppe,
Proto-Indo-European
Saturday, October 26, 2019
Warlike herders and their weapons
Who had the best gear? The Yamnaya guys? And if it came down to it, who would've won an all out rumble? Let me know your thoughts after reading this paper...
The weaponry of the pastoral societies in the context of the weaponry of the steppe/forest-steppe communities: 5000-2350 BCSee also...
Labels:
archeology,
axe,
battle,
Catacomb,
Corded Ware Culture,
Cucuteni-Trypilla,
dagger,
Eastern Europe,
Indo-European,
Pontic-Caspian steppe,
Usatovo culture,
Usatovo daggers,
warlike,
weaponry,
weapons,
Yamnaya
Wednesday, October 16, 2019
The Battle Axe people came from the steppe (Malmstrom et al. 2019)
It's been obvious for a while now that the Corded Ware culture (CWC) and its Scandinavian variant, the Battle Axe culture (BAC), originated on the Pontic-Caspian steppe. However, Malmstrom et al. drive the point home in a new open access paper at Proceedings B [LINK]. From the paper, emphasis is mine:
The Neolithic period is characterized by major cultural transformations and human migrations, with lasting effects across Europe. To understand the population dynamics in Neolithic Scandinavia and the Baltic Sea area, we investigate the genomes of individuals associated with the Battle Axe Culture (BAC), a Middle Neolithic complex in Scandinavia resembling the continental Corded Ware Culture (CWC). We sequenced 11 individuals (dated to 3330–1665 calibrated before common era (cal BCE)) from modern-day Sweden, Estonia, and Poland to 0.26–3.24× coverage. Three of the individuals were from CWC contexts and two from the central-Swedish BAC burial ‘Bergsgraven’. By analysing these genomes together with the previously published data, we show that the BAC represents a group different from other Neolithic populations in Scandinavia, revealing stratification among cultural groups. Similar to continental CWC, the BAC-associated individuals display ancestry from the Pontic–Caspian steppe herders, as well as smaller components originating from hunter–gatherers and Early Neolithic farmers. Thus, the steppe ancestry seen in these Scandinavian BAC individuals can be explained only by migration into Scandinavia. Furthermore, we highlight the reuse of megalithic tombs of the earlier Funnel Beaker Culture (FBC) by people related to BAC. The BAC groups likely mixed with resident middle Neolithic farmers (e.g. FBC) without substantial contributions from Neolithic foragers. ... By contrast, the CWC individuals from Obłaczkowo in Poland (poz44 and poz81) show an extremely high proportion of steppe ancestry (greater than 90%), which is different from the later CWC-associated individuals excavated in Pikutkowo (Poland) [23], but similar to some other CWC-associated individuals from Germany, Lithuania, and Latvia [2,8,31]. Interestingly, these individuals with a large fraction of steppe ancestry have typically been dated to more than 2600 BCE, making them among the earliest CWC individuals genetically investigated. This observation, i.e. early CWC individuals resembled (genetically) Yamnaya-associated individuals, while later CWC groups show higher levels of European Neolithic farmer ancestry (Pearson's correlation coefficient: −0.51, p = 0.006) (figure 2), suggests an initial dispersal that occurred rapidly.See also...
Saturday, October 12, 2019
The Balkan connection
The hot topic at the moment is social inequality in Bronze Age Europe, thanks to a new paper by Mittnik et al. at Science. The full article is sitting behind an exceedingly robust paywall here.
However, the genotype dataset from the paper is freely available at the Max Planck Society's Edmond data repository here. Below is my Principal Component Analysis (PCA) of ancient West Eurasian genetic variation featuring 41 of the highest quality ancients from the new dataset. Almost all of them are from the Lech Valley in the Bavarian Alps, covering the period from the Bell Beaker culture (BBC) to the Middle Bronze Age (MBA). Two of the samples are from a mass Corded Ware culture (CWC) burial in the more northerly Tauber Valley.
I've also highlighted other ancients on the plot associated with the BBC and CWC from present-day Netherlands and Germany, respectively. The relevant PCA datasheet can be downloaded here.
Social stratification in ancient Europe is a fascinating topic, and it's an issue that I've started looking at myself (see here). However, I can't see any correlation between the inferred social standing of the individuals from the Lech and Tauber valleys and their positions in my PCA.
Nevertheless, the PCA is interesting in that it highlights considerable genetic heterogeneity within the Lech Valley BBC population. Indeed, how is this heterogeneity even possible, if, as per Mittnik et al., ancient DNA "has shown that the spread of the BBC throughout continental Europe did not involve large-scale migrations"?
Below is another version of my PCA, but this time focusing on three males: Lech Valley Beakers UNTA58_68Sk1 and WEHR_1192SkA, as well as ALT_4 from the aforementioned mass CWC grave in the Tauber Valley. Note that UNTA58_68Sk1 and WEHR_1192SkA represent genetically the most southern and northern, respectively, Lech Valley BBC samples that had enough data to be run in my analysis. I chose to focus on males because they carry the Y-chromosome, which can be informative about male-mediated ancient population expansions.
The PCA outcomes for these individuals are generally in line with their results in other types of genetic analyses, including those based on formal statistics. For instance, compared to the other two, ALT_4 harbors excess early steppe herder ancestry, UNTA58_68Sk1 excess early European farmer ancestry, and WEHR_1192SkA excess European hunter-gatherer ancestry. Moreover...
- UNTA58_68Sk1 shows a non-local isotopic signature and belongs to Y-haplogroup G2a, a marker essentially missing from BBC populations north of the Alps, and is best modeled as a two-way mixture between Bronze Age populations from the Balkans and the Pontic-Caspian steppe (see here), which probably means that he was a migrant to the Lech Valley from south of the Alps - importantly, UNTA58_68Sk1 is not an isolated case, at least in the sense that several other BBC individuals from Bavaria, Bohemia, Hungary and Poland show varying ratios of Balkan-related ancestry, although almost all of these people are women - WEHR_1192SkA is very similar to Bell Beakers from the northern Netherlands with whom he shares the R1b-P312 Y-haplogroup, suggesting that he was part of a population that moved into the Lech Valley from potentially as far away as the North Sea coast - although ALT_4 probably shares the R1b-L51 Y-haplogroup with WEHR_1192SkA and many other BBC and Bronze Age individuals from the Bavarian Alps and surrounds, this can't be used as evidence of significant local genetic continuity after the CWC period, especially considering the comparatively eastern genome-wide structure of ALT_4.Of course, archeological data suggest that the BBC was influenced in some important ways by the Copper and Bronze Age cultures of the Balkans and Carpathian Basin. So much so, in fact, that Marija Gimbutas, author of The Civilization of the Goddess, believed that the BBC originated in the Balkans from a synthesis of the local Vucedol culture and the intrusive Yamnaya culture from the Pontic-Caspian steppe. Considering the ancient DNA evidence, however, the main demographic center of the early BBC could not have been south of the Alps. Rather, it appears that early BBC and even CWC groups from north of the Alps moved into the Balkans and Carpathian Basin, where they may have established contacts with the local elites. If so, this might explain the significant southern cultural influences on the BBC, but limited accompanying genetic impact. This scenario also has support from archeological data (for instance, see here). See also... Is Yamnaya overrated? The Boscombe Bowmen Single Grave > Bell Beakers
Thursday, September 26, 2019
Is Yamnaya overrated?
Four years after the publication of the seminal ancient DNA paper Massive migration from the steppe is a source for Indo-European languages in Europe by Haak et al., we're still waiting for some of its loose ends to be finally tied up with new samples. In particular...
- if the men of the Corded Ware culture (CWC) were, by and large, derived from the population of the Yamnaya culture, then where are the Yamnaya samples with R1a-M417, the main CWC Y-haplogroup? - if the men of the Bell Beaker culture (BBC) were also, by and large, derived from the population of the Yamnaya culture, then where are the Yamnaya samples with R1b-P312, the main BBC Y-haplogroup? - and, most crucially, if R1b-L51, which includes R1b-P312, and is nowadays by far the most important Y-haplogroup in Western Europe, arrived there from the Pontic-Caspian steppe, then why hasn't it yet appeared in any of the ancient DNA from this part of Eastern Europe or surrounds, except of course in samples that are too young to be relevant?I'm certainly not suggesting that, in hindsight, the said paper now looks fundamentally flawed. In fact, I'd say that it has aged remarkably well, especially considering how fast things are moving in the field of ancient genomics. But those loose ends really need tying up, one way or another. It's now time. So someone out there, please, let us know finally if you have the relevant Yamnaya samples. And if you don't, that's OK too, just tell us what you do have. Indeed, it'd be nice know a few basic details about the thousands of samples that have been successfully sequenced in various labs and are waiting to be published. A lot of people would appreciate it. See also... Corded Ware as an offshoot of Hungarian Yamnaya (Anthony 2017) Hungarian Yamnaya > Bell Beakers? Late PIE ground zero now obvious; location of PIE homeland still uncertain, but...
Wednesday, September 11, 2019
Y-haplogroup R1a and mental health
I've updated my map of pre-Corded Ware culture R1a samples with a couple of new entries from Central and South Asia (the original is still here). However, before any of you get overly excited, please note that these samples aren't older than the Corded Ware culture. The reason I added them to my map is to counter the ongoing absurd claims online that South Asian R1a isn't derived from European R1a.
Just in case the map can't be viewed in all of its glory in some devices, here's what the fine print says:
The oldest example of R1a in ancient DNA from Central Asia is dated to 2132-1940 calBCE (ID I3770, Narasimhan 2019). Moreover, this sequence is closely related to much older R1a samples from Central, Eastern and Northern Europe, and phylogenetically nested within their diversity. Thus, it must surely represent a population expansion from Europe to Central Asia. Indeed, it's also associated with the Bronze Age Andronovo archeological culture, which is usually seen as an offshoot of the Corded Ware culture (CWC) of Late Neolithic Europe. The vast majority of present-day R1a lineages in Central Asia are closely related to that of I3770, and so must also ultimately derive from Europe. The oldest instance of R1a in ancient DNA from South Asia is dated to just 1044-922 calBCE (ID I12457, Narasimhan 2019). This sequence, as well as the vast majority of present-day South Asian R1a lineages, are closely related to much older R1a samples from Central, Eastern and Northern Europe, and phylogenetically nested within their diversity. Thus, they must surely represent a population expansion from Europe to South Asia via Central Asia, in all likelihood during the Bronze Age. Even if R1a existed in South Asia before the Bronze Age, which is extremely unlikely, because it's found in samples from indigenous European hunter-gatherers, the vast majority of present-day R1a lineages in South Asia must be ultimately from Europe.The idea that most, if not all, South Asian R1a is derived from European R1a seriously scares a lot of people. This is obvious in many online discussions on the topic. I suspect they're so frightened by it because, in their minds, it has the potential to encourage discrimination and even racism, perhaps by re-defining the colonization of much of the world by European nations in the recent past as the natural order of things? In any case, clearly we're dealing with some sort of mass phobia here. I've got advice for those of you suffering from this problem: if you're honestly worried that the geographic provenance and expansion history of some Y-haplogroup is going to negatively impact on your life in any meaningful way, then it's time to find yourself a quality mental health professional. All the best with that. See also... The mystery of the Sintashta people The Poltavka outlier Yamnaya isn't from Iran just like R1a isn't from India
Labels:
ancient DNA,
Central Asia,
Corded Ware Culture,
Eastern Europe,
Eurasia,
India,
Indo-Aryan,
Indo-Iranian,
oldest R1a,
paternal ancestry,
Pontic-Caspian steppe,
R1a,
R1a origin,
R1a-M417,
R1a-Z645,
R1a-Z93,
South Asia
Thursday, September 5, 2019
On the surprising genetic origins of the Harappan people (Shinde et al. 2019)
The long awaited paper with ancient DNA from the Indus Valley Civilization (IVC) site of Rakhigarhi has finally arrived. Courtesy of Shinde et al. at Current Biology:
An ancient Harappan genome lacks ancestry from Steppe pastoralists or Iranian farmersThe bad news is that the paper features just one low coverage IVC genome, and it belongs to a female, so there's no Y-haplogroup. However, importantly, this individual is very similar to genetic outliers from Bronze Age West and Central Asia known as Indus_Periphery. So much so, in fact, that they could easily be from the same gene pool. This, of course, gives strong support to the idea that Indus_Periphery is a useful stand-in for the real IVC population (see here). Surprisingly, despite being largely of West Eurasian origin, the IVC people possibly didn't harbor any ancestry from the Neolithic farmers of the Fertile Crescent or even the Iranian Plateau. That's because, according to Shinde et al., their West Eurasian ancestors separated genetically from those of the early Holocene populations of what is now western and northern Iran around 12,000 BCE. In other words, well before the advent of agriculture. This surely complicates matters for those arguing that Indo-European languages may have arrived in the Indian subcontinent with early farmers via the Iranian Plateau. The more widely accepted theory is that Indo-European languages spread into South Asia with Bronze Age pastoralists from the Eurasian steppes. See here... Update 05/09/2019: I had a quick look at the ancient Rakhigarhi individual with qpAdm, just to confirm for myself that she was indeed largely of West Eurasian origin and practically indistinguishable from Indus_Periphery. The genotype data that I used are freely available here.
IND_Rakhigarhi_BA IRN_Ganj_Dareh_N 0.711±0.065 Onge 0.232±0.067 RUS_Tyumen_HG 0.057±0.059 chisq 13.251 tail prob 0.0392147 Full output Indus_Periphery IRN_Ganj_Dareh_N 0.674±0.015 Onge 0.237±0.014 RUS_Tyumen_HG 0.090±0.012 chisq 14.877 tail prob 0.0212326 Full output Indus_Periphery IND_Rakhigarhi_BA 0.946±0.074 Onge 0.054±0.074 chisq 10.358 tail prob 0.169152 Full outputThis does appear to be the case, although it's also obvious that my models are missing something important because their statistical fits are rather poor. I'm guessing the main problem is trying to use the Onge people of the Andaman Islands as a proxy for the indigenous foragers of the Indian subcontinent. See also... Y-haplogroup R1a and mental health
Labels:
Andronovo,
Dravidian,
Eurasian steppe,
farming,
Harappa,
India,
Indo-Aryan,
Indo-European,
Indo-Iranian,
Indus Valley Civilization,
Iranian Plateau,
IVC,
R1a-M417,
R1a-Z93,
Rakhigarhi,
Sintashta,
South Asia
Monday, September 2, 2019
Commoner or elite?
I recently started looking at the correlations between Y-chromosome haplogroups and social standing in ancient Europe, and was surprised by what I learned about the five currently sampled prehistoric Scandinavians belonging to Y-haplogroup R1b. I certainly wasn't expecting to uncover these stories about a mass human sacrifice, a bog body, and an Arctic circle warrior:
- The earliest Scandinavian in the ancient DNA record belonging to R1b comes from a grave site in what is now northern Norway (VK531, Margaryan et al. 2019). This individual has a genome-wide profile similar to that of local Mesolithic hunter-gatherers, but is dated to just ~2,400 BCE. During this time, Scandinavia was dominated by a "new" population associated with the Battle-Axe culture (BAC), with high levels of ancestry from the steppes of Eastern Europe. Since VK531 wasn't buried with any BAC grave goods, and indeed with no grave goods at all, it's possible that he may have been from a remnant forager population that was displaced and ultimately forced into extinction. - R1b-U106 is today by far the most common R1b subclade in Scandinavia, but it's not yet clear how it managed to attain this status. Was it perhaps through elite dominance? The earliest ancient individual belonging to R1b-U106 is dated to 2275-2032 calBCE and comes from a Late Neolithic, likely post-BAC burial ground in what is now Sweden (RISE98, Lilla Beddinge, grave 49, southern skeleton, Allentoft et al. 2015). However, RISE98 wasn't buried in any way that would suggest he was an individual of high social standing. In fact, he was found in a mass grave, along with two other adults and two infants, possibly representing a human sacrifice. The only artefact in the grave was a bone needle. More details are available here. - During the Nordic Bronze Age it became customary for Scandinavian elites to be laid to rest in richly furnished barrows, while commoners were buried in flat graves with few or no offerings. Human remains recovered from a "commoner" flat grave cemetery dated to the Early Bronze Age near the present-day city of Aalborg, northern Denmark, included the skeleton of a male belonging to Y-haplogroup R1b-M269 (RISE47, grave 3, skeleton 8, Allentoft et al. 2015). Keep in mind, however, that this might have been another case of an ancient Scandinavian R1b-U106 if not for missing data. A flint dagger was found alongside one of the skeletons in this cemetery, but RISE47 wasn't accompanied by any grave goods (see here). - One of the most amazing archeological discoveries made in Scandinavia is the Trundholm Sun Chariot. Found in a peat bog on the island of Zealand, Denmark, in 1902, it's thought to be an Indo-European religious artefact dating back to the Nordic Bronze Age; a representation of a horse pulling the sun and perhaps also the moon in a spoked wheel chariot. Another important discovery in a peat bog near Trundholm dating to the Nordic Bronze Age was the body of a man belonging to R1b-M269 (RISE276, Trundholm mose II, bog find 1940, Allentoft et al. 2015). However, chances are slim that RISE276 was a charioteer or, say, a spiritual guru who accidentally drowned in the bog. Most Danish bog bodies are thought to have belonged to sacrificial victims or executed criminals. - Interestingly, the earliest likely Scandinavian warrior belonging to R1b, and also R1b-U106, is from an early Iron Age burial in present-day northwestern Norway (VK418, Margaryan et al. 2019). This site isn't quite as far north as the grave of the above mentioned VK531, but it's still well within the Arctic circle. Apparently, VK418 was buried with some impressive weapons, potentially of "eastern origin", including a shield, spearheads and a sword. Who knows, he may even have been an elite warrior for his time and place?The other two main Scandinavian Y-haplogroups, I1a and R1a, haven't yet been found in prehistoric Nordic remains from such, shall we say, depressing burials. That's not to say, of course, that they won't be sooner or later. RISE175, from Allentoft et al. 2015, is currently the only individual who fits the bill as a representative of the Nordic Bronze Age elite. He was buried in a barrow grave in what is now southwest Sweden and probably belongs to Y-haplogroup I1a. That's not much to go on, but perhaps it's a sign of things to come? See also... Isotopes vs ancient DNA in prehistoric Scandinavia Who were the people of the Nordic Bronze Age? They came, they saw, and they mixed
Labels:
ancient DNA,
barrow,
Battle-Axe culture,
commoner,
Corded Ware Culture,
Denmark,
elite,
flat grave,
Germanic,
I1,
Indo-European,
Nordic Bronze Age,
R1a-M417,
R1a-Z284,
R1b,
R1b-M269,
R1b-U106,
Scandinavia,
Sweden
Tuesday, August 27, 2019
Isotopes vs ancient DNA in prehistoric Scandinavia
Four of the samples from the recent Frei et al. paper on human mobility in prehistoric southern Scandinavia are in my Global25 datasheets. Their genomes were published along with Allentoft et al. back in 2015. So I thought it might be interesting to check whether their strontium isotope ratios correlated with their genomic profiles.
In the Principal Component Analysis (PCA) below, RISE61 is a subtle outlier along the horizontal axis compared to the other three Nordic ancients, as well as a Danish individual representative of the present-day Danish gene pool. Also note that RISE61 shows the most unusual strontium isotope ratio (0.712588). The PCA was run with an online tool freely available here.
To help drive the point home, here's a figure from Frei et al., edited by me to show the positions of RISE47, RISE61 and RISE71. If RISE276 was also in this graph, he'd be sitting well under the "local" baseline, in roughly the same spot along the vertical axis as RISE47.
Interestingly, RISE61 belongs to Y-chromosome haplogroup R1a-M417, while RISE47 and RISE276, who appear to have been locals, both belong to R1b-M269. My guess is that RISE61 was a recent migrant from a more northerly part of Scandinavia dominated by the Battle-Axe culture (BAC). The BAC population was probably rich in R1a-M417 because it moved into Scandinavia from the Pontic-Caspian steppe via the East Baltic. This is what Frei et al. say about RISE61 and his burial site:
The double passage grave of Kyndeløse (Fig 1, S1 File) located on the island of Zealand yielded 70 individuals as well as a large number of grave goods, including flint artefacts, ceramics, and tooth and amber beads. We conducted strontium isotope analyses of seven individuals from Kyndeløse encompassing a period of c. 1000 years, indicating the prolonged use of this passage grave. The oldest of the seven individuals is a female (RISE 65) from whom we measured a “local” strontium isotope signature ( 87 Sr/ 86 Sr = 0.7099). Similar values were measured in five other individuals, including adult males and females. Only a single individual from Kyndeløse, an adult male (RISE 61) yielded a somewhat different strontium isotope signature of 87 Sr/ 86 Sr = 0.7126 which seems to indicate a non-local provenance. The skull of this male individual revealed healed porosities in the eye orbits, cribra orbitalia, a condition which is possibly linked to a vitamin deficiency during childhood, such as iron deficiency.By the way, RISE47 was buried in a flat grave, which suggests that he was a commoner. RISE276 was found in a peat bog in Trundholm, where the famous Trundholm sun chariot was discovered (see here). He may have been a human sacrifice. Citation... Frei KM, Bergerbrant S, Sjögren K-G, Jørkov ML, Lynnerup N, Harvig L, et al. (2019) Mapping human mobility during the third and second millennia BC in present-day Denmark. PLoS ONE 14(8): e0219850. https://doi.org/10.1371/journal.pone.0219850 See also... Commoner or elite? Who were the people of the Nordic Bronze Age? They came, they saw, and they mixed
Tuesday, August 20, 2019
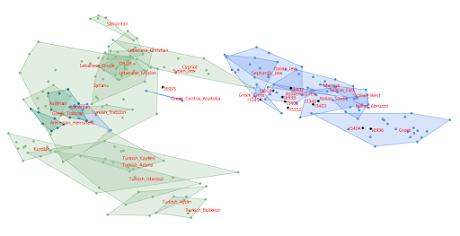
Roopkund Lake dead
Fifteen of the Roopkund Lake samples from the Harney et al. paper published today at Nature Communications made it into the Global25 datasheets. Look for the prefix IND_Roopkund here...
Global25 datasheet ancient scaled Global25 pop averages ancient scaled Global25 datasheet ancient Global25 pop averages ancientTheir genotypes are freely available in a ~590K SNP dataset via the Reich Lab here. I might be able to run more of the samples at some point if and when they're released in a dataset with more SNPs. In any case, much like everyone else, I don't have a clue how those Mediterranean migrants ended up in the Himalayas back in the 1800s, but I do know where they came from. Most appear to have been from Crete, while others from mainland Greece. However, one of the individuals that I was able to analyze with the Global25 was almost certainly an Anatolian Greek. Below are a couple of Principal Component Analyses (PCA) based on the Global25 data. The relevant datasheet is available here. I don't yet have a strong opinion about the origins of the earlier, typically South Asian Roopkund dead. They may have been visitors from all over India, or members of different castes from northern India. A PCA with six of these individuals can be seen here and the relevant datasheet gotten here. Any thoughts? Feel free to share them in the comments below. Update 23/08/2019: A new ~1240K SNP genotype dataset with the Roopkund Lake samples is now available here. More markers means that I can produce more accurate PCA and run almost twice as many of the samples. I've updated all of the datasheets accordingly. The links are the same. See also... Getting the most out of the Global25 A surprising twist to the Shirenzigou nomads story The Poltavka outlier
Subscribe to:
Posts (Atom)